Difference between revisions of "Timeline of DNA sequencing"
(→Numerical and visual data) |
|||
| (24 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
This is a timeline of [[wikipedia:DNA sequencing|DNA sequencing]], focusing on scientific developments. Major discoveries concerning DNA are exposed as background information. | This is a timeline of [[wikipedia:DNA sequencing|DNA sequencing]], focusing on scientific developments. Major discoveries concerning DNA are exposed as background information. | ||
| + | |||
| + | == Big Picture == | ||
{| class="sortable wikitable" | {| class="sortable wikitable" | ||
! Period !! Key developments | ! Period !! Key developments | ||
|- | |- | ||
| − | | | + | | 1930s–1940s || [[wikipedia:Molecular biology|Molecular biology]] originates.<ref name="Molecular Biology Stanford"/> |
|- | |- | ||
| − | | | + | | 1950s–1960s || Period of extraordinary development in molecular biology, which becomes institutionalized. The structure of DNA is uncovered by [[wikipedia:James Watson|James Watson]] and [[wikipedia:Francis Crick|Francis Crick]]. [[wikipedia:Frederick Sanger|Frederick Sanger]] plays a seminal role in the creation of influential DNA sequencing techniques in the 1950s and 1960s.<ref>{{cite web|title=Sequencing the worm|url=http://www.yourgenome.org/stories/sequencing-the-worm|website=yourgenome.org|accessdate=1 April 2017}}</ref><ref name="Molecular Biology Stanford">{{cite web|title=Molecular Biology|url=https://plato.stanford.edu/entries/molecular-biology/|website=stanford.edu|accessdate=1 April 2017}}</ref> |
|- | |- | ||
| − | | 1970s || First-generation sequencing technologies merge, including the [[wikipedia:Maxam–Gilbert sequencing|Maxam-Gilbert method]], and the [[wikipedia:Sanger sequencing|Sanger method.<ref>{{cite web|title=DNA sequencing|url=https://www.britannica.com/science/DNA-sequencing|website=britannica.com|accessdate=1 April 2017}}</ref> [[wikipedia:Molecular biology|Molecular biology]] goes [[wikipedia:genomic|genomic]].<ref name="Molecular Biology Stanford"/> | + | | 1970s || First-generation sequencing technologies merge, including the [[wikipedia:Maxam–Gilbert sequencing|Maxam-Gilbert method]], and the [[wikipedia:Sanger sequencing|Sanger method]].<ref>{{cite web|title=DNA sequencing|url=https://www.britannica.com/science/DNA-sequencing|website=britannica.com|accessdate=1 April 2017}}</ref> [[wikipedia:Molecular biology|Molecular biology]] goes [[wikipedia:genomic|genomic]].<ref name="Molecular Biology Stanford"/> |
|- | |- | ||
| − | | | + | | 1980s–1990s || After the development of sequencing techniques, efforts are focused in sequencing the human genome, giving rise to the [[wikipedia:Human Genome Project|Human Genome Project]]. Scientists start sequencing DNA using capillary sequencers.<ref name="Molecular Biology Stanford"/><ref>{{cite web|title=What is capillary sequencing?|url=http://www.yourgenome.org/facts/what-is-capillary-sequencing|website=yourgenome.org|accessdate=1 April 2017}}</ref> |
|- | |- | ||
| 2000s || Second generation of DNA sequencing and RNA sequencing launches.<ref>{{cite book|title=Biotechnology|edition=Ulrich Kück, Nicole Frankenberg-Dinkel|url=https://books.google.com.ar/books?id=65ynCgAAQBAJ&pg=PA198&lpg=PA198&dq=%22DNA+sequencing%22+%222000s%22&source=bl&ots=5OI-Z_zAJx&sig=bhMYDkhC39CsYEboJQR0uXY-Hwc&hl=en&sa=X&ved=0ahUKEwjap4qEgoLTAhWMh5AKHfHaB8cQ6AEIKTAC#v=onepage&q=%22DNA%20sequencing%22%20%222000s%22&f=false|accessdate=1 April 2017}}</ref> | | 2000s || Second generation of DNA sequencing and RNA sequencing launches.<ref>{{cite book|title=Biotechnology|edition=Ulrich Kück, Nicole Frankenberg-Dinkel|url=https://books.google.com.ar/books?id=65ynCgAAQBAJ&pg=PA198&lpg=PA198&dq=%22DNA+sequencing%22+%222000s%22&source=bl&ots=5OI-Z_zAJx&sig=bhMYDkhC39CsYEboJQR0uXY-Hwc&hl=en&sa=X&ved=0ahUKEwjap4qEgoLTAhWMh5AKHfHaB8cQ6AEIKTAC#v=onepage&q=%22DNA%20sequencing%22%20%222000s%22&f=false|accessdate=1 April 2017}}</ref> | ||
|- | |- | ||
|} | |} | ||
| + | |||
| + | == Numerical and visual data == | ||
| + | |||
| + | === Google Scholar === | ||
| + | |||
| + | The following table summarizes per-year mentions on Google Scholar as of May 22, 2021. | ||
| + | |||
| + | {| class="sortable wikitable" | ||
| + | ! Year | ||
| + | ! dna sequencing | ||
| + | ! next generation dna sequencing | ||
| + | ! nanopore dna sequencing | ||
| + | ! automated dna sequencing | ||
| + | ! high-throughput dna sequencing | ||
| + | ! electrophoresis dna sequencing | ||
| + | |- | ||
| + | | 1980 || 1,660 || 164 || || 100 || 11 || 4,710 | ||
| + | |- | ||
| + | | 1985 || 5,880 || 475 || 1 || 361 || 20 || 10,300 | ||
| + | |- | ||
| + | | 1990 || 15,900 || 1,490 || || 1,4980 || 66 || 19,400 | ||
| + | |- | ||
| + | | 1995 || 58,000 || 3,340 || 5 || 4,320 || 369 || 26,800 | ||
| + | |- | ||
| + | | 2000 || 106,000 || 10,800 || 62 || 11,500 || 3,880 || 46,000 | ||
| + | |- | ||
| + | | 2002 || 115,000 || 14,100 || 129 || 12,900 || 7,290 || 44,000 | ||
| + | |- | ||
| + | | 2004 || 125,000 || 18,200 || 295 || 14,300 || 11,600 || 47,200 | ||
| + | |- | ||
| + | | 2006 || 128,000 || 19,800 || 477 || 14,200 || 15,700 || 48,900 | ||
| + | |- | ||
| + | | 2008 || 126,000 || 24,500 || 689 || 15,100 || 18,400 || 50,500 | ||
| + | |- | ||
| + | | 2010 || 135,000 || 35,300 || 1,140 || 16,600 || 23,100 || 51,800 | ||
| + | |- | ||
| + | | 2012 || 171,000 || 60,800 || 1,780 || 21,500 || 34,500 || 57,300 | ||
| + | |- | ||
| + | | 2014 || 174,000 || 76,900 || 2,100 || 21,900 || 48,500 || 55,700 | ||
| + | |- | ||
| + | | 2016 || 161,000 || 81,200 || 2,770 || 24,400 || 57,800 || 51,900 | ||
| + | |- | ||
| + | | 2017 || 141,000 || 79,800 || 3,630 || 25,900 || 57,500 || 49,700 | ||
| + | |- | ||
| + | | 2018 || 113,000 || 72,600 || 4,630 || 27,200 || 54,500 || 47,900 | ||
| + | |- | ||
| + | | 2019 || 84,000 || 57,600 || 5,790 || 28,600 || 48,800 || 44,800 | ||
| + | |- | ||
| + | | 2020 || 77,200 || 43,600 || 8,460 || 26,000 || 38,000 || 39,800 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | [[File:Dna tb.png|thumb|center|700px]] | ||
| + | |||
| + | === Google Trends === | ||
| + | The comparative chart below shows {{w|Google Trends}} data for DNA sequencing (Search Term) and DNA sequencing (Topic), from January 2004 to February 2021, when the screenshot was taken. Interest in the topic is also ranked by country and displayed on world map.<ref>{{cite web |title=DNA sequencing |url=https://trends.google.com/trends/explore?date=all&q=DNA%20sequencing,%2Fm%2F04c45l |website=Google Trends |access-date=15 February 2021}}</ref> | ||
| + | |||
| + | [[File:DNA sequencing gt.jpg|thumb|center|600px]] | ||
| + | |||
| + | === Google Ngram Viewer === | ||
| + | |||
| + | The chart below shows {{w|Google Ngram Viewer}} data for DNA sequencing, from 1950 to 2019.<ref>{{cite web |title=DNA sequencing |url=https://books.google.com/ngrams/graph?content=DNA+sequencing&year_start=1950&year_end=2019&corpus=26&smoothing=3&case_insensitive=true |website=books.google.com |access-date=16 February 2021 |language=en}}</ref> | ||
| + | |||
| + | [[File:DNA sequencing ngram.jpg|thumb|center|700px]] | ||
| + | |||
| + | === Wikipedia Views === | ||
| + | |||
| + | The chart below shows pageviews of the English Wikipedia article {{w|DNA sequencing}}, on desktop, mobile-web, desktop-spider, mobile-web-spider and mobile app, from July 2015; to January 2021.<ref>{{cite web |title=DNA sequencing |url=https://wikipediaviews.org/displayviewsformultiplemonths.php?page=DNA+sequencing&allmonths=allmonths-api&language=en&drilldown=all |website=wikipediaviews.org |access-date=21 February 2021}}</ref> | ||
| + | |||
| + | [[File:DNA sequencing wv.jpg|thumb|center|400px]] | ||
| + | |||
| + | === Other === | ||
<gallery align="center" perrow="2" widths="400" heights="400" mode="nolines"> | <gallery align="center" perrow="2" widths="400" heights="400" mode="nolines"> | ||
| Line 27: | Line 101: | ||
</gallery> | </gallery> | ||
| − | ==Full timeline== | + | ==Full timeline== |
{| class="sortable wikitable" | {| class="sortable wikitable" | ||
! Year/period !! Type of event !! Event !! Location | ! Year/period !! Type of event !! Event !! Location | ||
|- | |- | ||
| − | | | + | | 1869–1871 || Scientific development || Swiss physician [[wikipedia:Friedrich Miescher|Friedrich Miescher]] first discovers and isolates Deoxyribonucleic acid (DNA). In 1871 Miescher publishes paper identifying the presence of ''nuclein'' (now known as DNA) and associated proteins, in the [[wikipedia:cell nucleus|cell nucleus]].<ref>{{cite web|title=Friedrich Miescher and the discovery of DNA|url=http://www.sciencedirect.com/science/article/pii/S0012160604008231|website=sciencedirect.com|accessdate=29 March 2017}}</ref> || [[wikipedia:Germany|Germany]] ([[wikipedia:Tübingen|Tübingen]]) |
|- | |- | ||
| 1882 || Scientific development || German biochemist [[wikipedia:Albrecht Kossel|Albrecht Kossel]] differentiates [[wikipedia:ARN|ARN]] and [[wikipedia:DNA|DNA]].<ref name="Textbook of Biochemistry for Medical Students"/> || | | 1882 || Scientific development || German biochemist [[wikipedia:Albrecht Kossel|Albrecht Kossel]] differentiates [[wikipedia:ARN|ARN]] and [[wikipedia:DNA|DNA]].<ref name="Textbook of Biochemistry for Medical Students"/> || | ||
| Line 40: | Line 114: | ||
| 1902 || Scientific development || German biologist [[wikipedia:Theodor Boveri|Theodor Boveri]], working independently from [[wikipedia:Walter Sutton|Walter Sutton]], recognizes that the behavior of Mendel’s particles during the production of gametes in peas precisely parallels the behavior of chromosomes at meiosis, suggesting that [[wikipedia:chromosome|chromosome]]s could be shown to bear the material of heredity. This would further give rise to the so called [[wikipedia:Boveri–Sutton chromosome theory|Boveri–Sutton chromosome theory]], which is recognized as a fundamental unifying theory of genetics which identifies chromosomes as the carriers of genetic material.<ref name="An Introduction to Genetic Analysis. 7th edition.">{{cite book|title=An Introduction to Genetic Analysis. 7th edition.|url=https://www.ncbi.nlm.nih.gov/books/NBK22088/|accessdate=29 March 2017|chapter=Historical development of the chromosome theory}}</ref> || [[wikipedia:Germany|Germany]] | | 1902 || Scientific development || German biologist [[wikipedia:Theodor Boveri|Theodor Boveri]], working independently from [[wikipedia:Walter Sutton|Walter Sutton]], recognizes that the behavior of Mendel’s particles during the production of gametes in peas precisely parallels the behavior of chromosomes at meiosis, suggesting that [[wikipedia:chromosome|chromosome]]s could be shown to bear the material of heredity. This would further give rise to the so called [[wikipedia:Boveri–Sutton chromosome theory|Boveri–Sutton chromosome theory]], which is recognized as a fundamental unifying theory of genetics which identifies chromosomes as the carriers of genetic material.<ref name="An Introduction to Genetic Analysis. 7th edition.">{{cite book|title=An Introduction to Genetic Analysis. 7th edition.|url=https://www.ncbi.nlm.nih.gov/books/NBK22088/|accessdate=29 March 2017|chapter=Historical development of the chromosome theory}}</ref> || [[wikipedia:Germany|Germany]] | ||
|- | |- | ||
| − | | 1906 || Scientific development || German biochemist [[wikipedia:Albrecht Kossel|Albrecht Kossel]] describes the four bases in nucleic acids (denine, cytosine, guanine, thymine). In 1910 Kossel would be awarded a Nobel Prize in Physiology or Medicine for that work.<ref name="Textbook of Biochemistry for Medical Students">{{cite book|last1=Vasudevan|first1=D M|last2=Sreekumari|first2=S|last3=Vaidyanathan|first3=Kannan|title=Textbook of Biochemistry for Medical Students|url=https://books.google.com.ar/books?id=nQz8AAAAQBAJ&pg=PA559&lpg=PA559&dq=%22Albrecht+Kossel+%22+%22nucleotides%22&source=bl&ots=--F4E8lG5t&sig=FiFTSlbO4tKwrTxYeUTC3tEDY9c&hl=en&sa=X&ved=0ahUKEwiby4qXu_zSAhUFFZAKHUEGArgQ6AEILTAF#v=onepage&q=%22Albrecht%20Kossel%20%22%20%22nucleotides%22&f=false|accessdate=29 March 2017}}</ref> || | + | | 1906 || Scientific development || German biochemist [[wikipedia:Albrecht Kossel|Albrecht Kossel]] describes the four bases in nucleic acids (denine, cytosine, guanine, thymine). In 1910 Kossel would be awarded a Nobel Prize in Physiology or Medicine for that work.<ref name="Textbook of Biochemistry for Medical Students">{{cite book|last1=Vasudevan|first1=D M|last2=Sreekumari|first2=S|last3=Vaidyanathan|first3=Kannan|title=Textbook of Biochemistry for Medical Students|url=https://books.google.com.ar/books?id=nQz8AAAAQBAJ&pg=PA559&lpg=PA559&dq=%22Albrecht+Kossel+%22+%22nucleotides%22&source=bl&ots=--F4E8lG5t&sig=FiFTSlbO4tKwrTxYeUTC3tEDY9c&hl=en&sa=X&ved=0ahUKEwiby4qXu_zSAhUFFZAKHUEGArgQ6AEILTAF#v=onepage&q=%22Albrecht%20Kossel%20%22%20%22nucleotides%22&f=false|accessdate=29 March 2017}}</ref> || |
|- | |- | ||
| 1944 || Scientific development || Canadian-American researchers [[wikipedia:Oswald Avery|Oswald Avery]] and [[wikipedia:Colin Munro MacLeod|Colin Munro MacLeod]], along with American geneticist [[wikipedia:Maclyn McCarty|Maclyn McCarty]], working on experiments on pneumococcal bacteria, establish that DNA could transform the properties of cells. As a result, a number of researchers would begin investigating the structure of DNA, hoping that this would reveal how the molecule works.<ref name="The path to sequencing nucleic acids"/> || | | 1944 || Scientific development || Canadian-American researchers [[wikipedia:Oswald Avery|Oswald Avery]] and [[wikipedia:Colin Munro MacLeod|Colin Munro MacLeod]], along with American geneticist [[wikipedia:Maclyn McCarty|Maclyn McCarty]], working on experiments on pneumococcal bacteria, establish that DNA could transform the properties of cells. As a result, a number of researchers would begin investigating the structure of DNA, hoping that this would reveal how the molecule works.<ref name="The path to sequencing nucleic acids"/> || | ||
| Line 46: | Line 120: | ||
| 1950 || Discovery || Austro-Hungarian biochemist [[wikipedia:Erwin Chargaff|Erwin Chargaff]] discovers that concentrations of thymine and adenine, and cytosine and guanine, are always found in equal amounts in samples of DNA. This suggests that A always pairs with T and C always pairs with G in a proportion that would later become known as Chargaff's ratio, an important clue for solving the structure of DNA. <ref>{{cite web|title=Chargaff's Ratio|url=http://www.hhmi.org/biointeractive/chargaffs-ratio|website=hhmi.org|accessdate=29 March 2017}}</ref> || | | 1950 || Discovery || Austro-Hungarian biochemist [[wikipedia:Erwin Chargaff|Erwin Chargaff]] discovers that concentrations of thymine and adenine, and cytosine and guanine, are always found in equal amounts in samples of DNA. This suggests that A always pairs with T and C always pairs with G in a proportion that would later become known as Chargaff's ratio, an important clue for solving the structure of DNA. <ref>{{cite web|title=Chargaff's Ratio|url=http://www.hhmi.org/biointeractive/chargaffs-ratio|website=hhmi.org|accessdate=29 March 2017}}</ref> || | ||
|- | |- | ||
| − | | 1952 || Scientific development || American bacteriologist [[wikipedia:Alfred Hershey|Alfred Hershey]] and American geneticist [[wikipedia:Martha Chase|Martha Chase]] conduct a series of experiments (today known as [[wikipedia:Hershey–Chase experiment|Hershey–Chase experiment]]) to demonstrate that DNA, rather than protein, carries our genetic information.<ref name="DNA Methylation: Molecular Biology and Biological Significance">{{cite book|last1=Saluz|first1=Jost, H.|title=DNA Methylation: Molecular Biology and Biological Significance|url=https://books.google.com.ar/books?id=ySfyBwAAQBAJ&pg=PA3&dq=%221952%22+%22+Hershey-Chase%22&hl=en&sa=X&ved=0ahUKEwir88m-x_zSAhUHhpAKHVX_A0AQ6AEIITAC#v=onepage&q=%221952%22%20%22%20Hershey-Chase%22&f=false|accessdate=29 March 2017}}</ref> || | + | | 1952 || Scientific development || American bacteriologist [[wikipedia:Alfred Hershey|Alfred Hershey]] and American geneticist [[wikipedia:Martha Chase|Martha Chase]] conduct a series of experiments (today known as [[wikipedia:Hershey–Chase experiment|Hershey–Chase experiment]]) to demonstrate that DNA, rather than protein, carries our genetic information.<ref name="DNA Methylation: Molecular Biology and Biological Significance">{{cite book|last1=Saluz|first1=Jost, H.|title=DNA Methylation: Molecular Biology and Biological Significance|url=https://books.google.com.ar/books?id=ySfyBwAAQBAJ&pg=PA3&dq=%221952%22+%22+Hershey-Chase%22&hl=en&sa=X&ved=0ahUKEwir88m-x_zSAhUHhpAKHVX_A0AQ6AEIITAC#v=onepage&q=%221952%22%20%22%20Hershey-Chase%22&f=false|accessdate=29 March 2017}}</ref> || |
|- | |- | ||
| 1952 || Scientific development || R. Markham and J. D. Smith publish paper reporting that the hydrolysis of RNA proceeds via a cyclic phosphate intermediate, which is then further hydrolysed to produce a nucleoside 2'-monophosphate or 3'-monophosphate.<ref name="Milestones of DNA technologies">{{cite web|title=Milestones of DNA technologies|url=http://www.nature.com/milestones/miledna/full/miledna01.html|publisher=Nature|accessdate=28 March 2017|doi=10.1038/nrg2250}}</ref> || | | 1952 || Scientific development || R. Markham and J. D. Smith publish paper reporting that the hydrolysis of RNA proceeds via a cyclic phosphate intermediate, which is then further hydrolysed to produce a nucleoside 2'-monophosphate or 3'-monophosphate.<ref name="Milestones of DNA technologies">{{cite web|title=Milestones of DNA technologies|url=http://www.nature.com/milestones/miledna/full/miledna01.html|publisher=Nature|accessdate=28 March 2017|doi=10.1038/nrg2250}}</ref> || | ||
| Line 58: | Line 132: | ||
| 1959 || Scientific development (non-human genome) || American molecular biologist [[wikipedia:Robert Sinsheimer|Robert Sinsheimer]] reports the genome of [[wikipedia:bacteriophage|bacteriophage]] [[wikipedia:Phi X 174|Phi X 174]] becoming the first DNA molecule purified to homogeneity.<ref name="DNA sequencing: bench to bedside and beyond"/> || | | 1959 || Scientific development (non-human genome) || American molecular biologist [[wikipedia:Robert Sinsheimer|Robert Sinsheimer]] reports the genome of [[wikipedia:bacteriophage|bacteriophage]] [[wikipedia:Phi X 174|Phi X 174]] becoming the first DNA molecule purified to homogeneity.<ref name="DNA sequencing: bench to bedside and beyond"/> || | ||
|- | |- | ||
| − | | | + | | 1961–1966 || Scientific development || American biochemist [[wikipedia:Marshall Warren Nirenberg|Marshall Warren Nirenberg]], working at the National Institute of Arthritic and Metabolic Diseases, discover the first "triplet" —a sequence of three bases of DNA that codes for one of the twenty amino acids that serve as the building blocks of proteins. By 1966, Nirenberg has deciphered over sixty triplets. Nirenberg's experimental system would be used, confirmed and extended by Indian-American biochemist [[wikipedia:Har Gobind Khorana|Har Gobind Khorana]], who, along with Nirenberg and [[wikipedia:Robert W. Holley|Robert W. Holley]], was be awarded the 1968 Nobel Prize in Physiology or Medicine "for their interpretation of the genetic code and its function in protein synthesis."<ref>{{cite web|title=Marshall Nirenberg (1927-) cracks the genetic code|url=http://www.genomenewsnetwork.org/resources/timeline/1961_Nirenberg.php|website=genomenewsnetwork.org|accessdate=30 March 2017}}</ref> || |
|- | |- | ||
| 1965 || Scientific development (non-human genome) || [[wikipedia:Escherichia coli|Escherichia coli]] [[wikipedia:alanine|alanine]] [[wikipedia:transfer RNA|transfer RNA]] becomes the first nucleic acid molecule to be sequenced by American biochemist [[wikipedia:Robert W. Holley|Robert W. Holley]] and colleagues.<ref name="DNA sequencing: bench to bedside and beyond"/><ref name="The sequence of sequencers: The history of sequencing DNA"/> || | | 1965 || Scientific development (non-human genome) || [[wikipedia:Escherichia coli|Escherichia coli]] [[wikipedia:alanine|alanine]] [[wikipedia:transfer RNA|transfer RNA]] becomes the first nucleic acid molecule to be sequenced by American biochemist [[wikipedia:Robert W. Holley|Robert W. Holley]] and colleagues.<ref name="DNA sequencing: bench to bedside and beyond"/><ref name="The sequence of sequencers: The history of sequencing DNA"/> || | ||
| Line 68: | Line 142: | ||
| 1972 || Scientific development || American microbbiologist [[wikipedia:Hamilton O. Smith|Hamilton O. Smith]], at [[wikipedia:Johns Hopkins University School of Medicine|Johns Hopkins University School of Medicine]], purifies the first site-specific "Type II" restriction enzyme, known as ''Hind II''. Restriction enzymes would enable scientists to identify base sequences and, in combination with other tools, to manipulate DNA —and genes by the way— as never before. Hamilton Smith is awarded the [[wikipedia:Nobel Prize in physiology or medicine|Nobel Prize in physiology or medicine]] in 1978.<ref>{{cite web|title=Hamilton O. Smith (1931-) discovers the first site-specific restriction enzyme|url=http://www.genomenewsnetwork.org/resources/timeline/1970_Smith.php|website=genomenewsnetwork.org|accessdate=30 March 2017}}</ref> || [[wikipedia:United States|United States]] | | 1972 || Scientific development || American microbbiologist [[wikipedia:Hamilton O. Smith|Hamilton O. Smith]], at [[wikipedia:Johns Hopkins University School of Medicine|Johns Hopkins University School of Medicine]], purifies the first site-specific "Type II" restriction enzyme, known as ''Hind II''. Restriction enzymes would enable scientists to identify base sequences and, in combination with other tools, to manipulate DNA —and genes by the way— as never before. Hamilton Smith is awarded the [[wikipedia:Nobel Prize in physiology or medicine|Nobel Prize in physiology or medicine]] in 1978.<ref>{{cite web|title=Hamilton O. Smith (1931-) discovers the first site-specific restriction enzyme|url=http://www.genomenewsnetwork.org/resources/timeline/1970_Smith.php|website=genomenewsnetwork.org|accessdate=30 March 2017}}</ref> || [[wikipedia:United States|United States]] | ||
|- | |- | ||
| − | | 1972 || Technology launch|| Using the 2-D fractionation method developed by [[wikipedia:Frederick Sanger|Frederick Sanger]], Belgian molecular biologist [[wikipedia:Walter Fiers|Walter Fiers]]' laboratory produces the first complete protein-coding gene sequence, that of the coat protein of bacteriophage MS2.<ref name="The sequence of sequencers: The history of sequencing DNA"/> || | + | | 1972 || Technology launch|| Using the 2-D fractionation method developed by [[wikipedia:Frederick Sanger|Frederick Sanger]], Belgian molecular biologist [[wikipedia:Walter Fiers|Walter Fiers]]' laboratory produces the first complete protein-coding gene sequence, that of the coat protein of bacteriophage MS2.<ref name="The sequence of sequencers: The history of sequencing DNA"/> || |
|- | |- | ||
| 1973 || Scientific development || American molecular biologists [[wikipedia:Walter Gilbert|Walter Gilbert]] and [[wikipedia:Allan Maxam|Allan Maxam]] publish first report of a nucleotide sequence with the lenght of 24 basepairs, using a method later known as [[wikipedia:wandering-spot analysis|wandering-spot analysis]].<ref name="Books on Google Play Synthetic Biology and Metabolic Engineering in Plants and Microbes Part B: Metabolism in Plants">{{cite book|title=Books on Google Play Synthetic Biology and Metabolic Engineering in Plants and Microbes Part B: Metabolism in Plants|url=https://books.google.com.ar/books?id=9jwADAAAQBAJ&pg=PA30&dq=%221973%22+%22Gilbert%22+and+%22Maxam%22+report+the+%22sequence%22+of+24&hl=en&sa=X&ved=0ahUKEwiPirvtnf_SAhVBkJAKHVG9D3AQ6AEIGDAA#v=onepage&q=%221973%22%20%22Gilbert%22%20and%20%22Maxam%22%20report%20the%20%22sequence%22%20of%2024&f=false|accessdate=30 March 2017}}</ref><ref name="Maxam-Gilbert">{{cite web|title=Maxam-Gilbert|url=https://binf.snipcademy.com/lessons/dna-sequencing-techniques/maxam-gilbert|website=snipcademy.com|accessdate=30 March 2017}}</ref><ref name="Genome Mapping: To Determine Disease Susceptibility"/> || | | 1973 || Scientific development || American molecular biologists [[wikipedia:Walter Gilbert|Walter Gilbert]] and [[wikipedia:Allan Maxam|Allan Maxam]] publish first report of a nucleotide sequence with the lenght of 24 basepairs, using a method later known as [[wikipedia:wandering-spot analysis|wandering-spot analysis]].<ref name="Books on Google Play Synthetic Biology and Metabolic Engineering in Plants and Microbes Part B: Metabolism in Plants">{{cite book|title=Books on Google Play Synthetic Biology and Metabolic Engineering in Plants and Microbes Part B: Metabolism in Plants|url=https://books.google.com.ar/books?id=9jwADAAAQBAJ&pg=PA30&dq=%221973%22+%22Gilbert%22+and+%22Maxam%22+report+the+%22sequence%22+of+24&hl=en&sa=X&ved=0ahUKEwiPirvtnf_SAhVBkJAKHVG9D3AQ6AEIGDAA#v=onepage&q=%221973%22%20%22Gilbert%22%20and%20%22Maxam%22%20report%20the%20%22sequence%22%20of%2024&f=false|accessdate=30 March 2017}}</ref><ref name="Maxam-Gilbert">{{cite web|title=Maxam-Gilbert|url=https://binf.snipcademy.com/lessons/dna-sequencing-techniques/maxam-gilbert|website=snipcademy.com|accessdate=30 March 2017}}</ref><ref name="Genome Mapping: To Determine Disease Susceptibility"/> || | ||
| Line 76: | Line 150: | ||
| 1976 || Scientific development (non-human genome) || Walter Fiers and coworkers at the [[wikipedia:University of Ghent|University of Ghent]] identify and publish the sequence of the first complete genome of Bacteriophage MS2.<ref name="Genome Mapping: To Determine Disease Susceptibility">{{cite book|last1=Schiller|first1=Jon|title=Genome Mapping: To Determine Disease Susceptibility|url=https://books.google.com.ar/books?id=cKOvgeg35C0C&pg=PA44&lpg=PA44&dq=%7C+1976+%7C%7C+%7C%7C+The+sequence+of+the+first+complete+genome+of+Bacteriophage+MS2+is+identified+and+published+by+Walter+Fiers+and+his+coworkers+at+the+%5B%5BUniversity+of+Ghent%5D%5D.&source=bl&ots=_6vWBl_wb-&sig=ZN8ksYYQPoIWimFHhWVxlMa5jr4&hl=en&sa=X&ved=0ahUKEwjqtouxsv_SAhXHPpAKHaAxBEIQ6AEIGjAA#v=onepage&q=%7C%201976%20%7C%7C%20%7C%7C%20The%20sequence%20of%20the%20first%20complete%20genome%20of%20Bacteriophage%20MS2%20is%20identified%20and%20published%20by%20Walter%20Fiers%20and%20his%20coworkers%20at%20the%20%5B%5BUniversity%20of%20Ghent%5D%5D.&f=false|accessdate=30 March 2017}}</ref> || | | 1976 || Scientific development (non-human genome) || Walter Fiers and coworkers at the [[wikipedia:University of Ghent|University of Ghent]] identify and publish the sequence of the first complete genome of Bacteriophage MS2.<ref name="Genome Mapping: To Determine Disease Susceptibility">{{cite book|last1=Schiller|first1=Jon|title=Genome Mapping: To Determine Disease Susceptibility|url=https://books.google.com.ar/books?id=cKOvgeg35C0C&pg=PA44&lpg=PA44&dq=%7C+1976+%7C%7C+%7C%7C+The+sequence+of+the+first+complete+genome+of+Bacteriophage+MS2+is+identified+and+published+by+Walter+Fiers+and+his+coworkers+at+the+%5B%5BUniversity+of+Ghent%5D%5D.&source=bl&ots=_6vWBl_wb-&sig=ZN8ksYYQPoIWimFHhWVxlMa5jr4&hl=en&sa=X&ved=0ahUKEwjqtouxsv_SAhXHPpAKHaAxBEIQ6AEIGjAA#v=onepage&q=%7C%201976%20%7C%7C%20%7C%7C%20The%20sequence%20of%20the%20first%20complete%20genome%20of%20Bacteriophage%20MS2%20is%20identified%20and%20published%20by%20Walter%20Fiers%20and%20his%20coworkers%20at%20the%20%5B%5BUniversity%20of%20Ghent%5D%5D.&f=false|accessdate=30 March 2017}}</ref> || | ||
|- | |- | ||
| − | | | + | | 1976–1977 || Technology launch|| [[wikipedia:Walter Gilbert|Walter Gilbert]] and [[wikipedia:Allan Maxam|Allan Maxam]] introduce their second method of DNA sequencing using chemical processes to terminate DNA strands. This method would be known as [[wikipedia:Maxam–Gilbert sequencing|Maxam–Gilbert sequencing]].<ref name="DNA sequencing: bench to bedside and beyond"/><ref name="Maxam-Gilbert"/> || |
|- | |- | ||
| 1977 || Technology launch|| [[wikipedia:Frederick Sanger|Frederick Sanger]] develops his "chain-termination" or dideoxy technique, which allows the use of chemical analogues of the deoxyribonucleotides (dNTPs) that are the monomers of DNA strands. This technique is considered a major breakthrough, having altered the progress of DNA sequencing.<ref name="The sequence of sequencers: The history of sequencing DNA"/> In 1980 Fred Sanger would be awarded his second Nobel Prize for Chemistry (this one along with Wally Gilbert and Paul Berg), for pioneering DNA sequencing methods.<ref name="Advanced Biology (sanger)"/> || | | 1977 || Technology launch|| [[wikipedia:Frederick Sanger|Frederick Sanger]] develops his "chain-termination" or dideoxy technique, which allows the use of chemical analogues of the deoxyribonucleotides (dNTPs) that are the monomers of DNA strands. This technique is considered a major breakthrough, having altered the progress of DNA sequencing.<ref name="The sequence of sequencers: The history of sequencing DNA"/> In 1980 Fred Sanger would be awarded his second Nobel Prize for Chemistry (this one along with Wally Gilbert and Paul Berg), for pioneering DNA sequencing methods.<ref name="Advanced Biology (sanger)"/> || | ||
| Line 82: | Line 156: | ||
| 1981 || Scientific development (non-human genome) || The genome of the [[wikipedia:cauliflower mosaic virus|cauliflower mosaic virus]] is first to be sequenced by [[wikipedia:shotgun sequencing|shotgun sequencing]].<ref>{{Cite journal|last=Gardner|first=Richard C.|last2=Howarth|first2=Alan J.|last3=Hahn|first3=Peter|last4=Brown-Luedi|first4=Marianne|last5=Shepherd|first5=Robert J.|last6=Messing|first6=Joachim|date=1981-06-25|title=The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing|url=http://nar.oxfordjournals.org/content/9/12/2871|journal=Nucleic Acids Research|language=en|volume=9|issue=12|pages=2871–2888|doi=10.1093/nar/9.12.2871|issn=0305-1048|pmid=6269062|pmc=326899|accessdate=30 March 2017}}</ref> | | 1981 || Scientific development (non-human genome) || The genome of the [[wikipedia:cauliflower mosaic virus|cauliflower mosaic virus]] is first to be sequenced by [[wikipedia:shotgun sequencing|shotgun sequencing]].<ref>{{Cite journal|last=Gardner|first=Richard C.|last2=Howarth|first2=Alan J.|last3=Hahn|first3=Peter|last4=Brown-Luedi|first4=Marianne|last5=Shepherd|first5=Robert J.|last6=Messing|first6=Joachim|date=1981-06-25|title=The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing|url=http://nar.oxfordjournals.org/content/9/12/2871|journal=Nucleic Acids Research|language=en|volume=9|issue=12|pages=2871–2888|doi=10.1093/nar/9.12.2871|issn=0305-1048|pmid=6269062|pmc=326899|accessdate=30 March 2017}}</ref> | ||
|- | |- | ||
| − | | 1983 || Technology launch|| American biochemist [[wikipedia:Kary Mullis|Kary Mullis]] develops the [[wikipedia:Polymerase chain reaction|Polymerase chain reaction]] (PCR), a technique used in molecular biology to rapidly amplify DNA. The technique would prove revolutionary, and Mullis would be awarded the 1993 [[wikipedia:Nobel Prize in Chemistry|Nobel Prize in Chemistry]]. <ref>{{cite book|last1=Clark|first1=David P.|title=Molecular Biology|url=https://books.google.com.ar/books?id=m1SmtB4CiywC&pg=PT655&dq=%221983%22+%22mullis%22&hl=en&sa=X&ved=0ahUKEwiHmMC-0P3SAhXEjJAKHWV7CDsQ6AEIHjAB#v=onepage&q=%221983%22%20%22mullis%22&f=false|accessdate=30 March 2017}}</ref> || [[wikipedia:United States|United States]] | + | | 1983 || Technology launch|| American biochemist [[wikipedia:Kary Mullis|Kary Mullis]] develops the [[wikipedia:Polymerase chain reaction|Polymerase chain reaction]] (PCR), a technique used in molecular biology to rapidly amplify DNA. The technique would prove revolutionary, and Mullis would be awarded the 1993 [[wikipedia:Nobel Prize in Chemistry|Nobel Prize in Chemistry]]. <ref>{{cite book|last1=Clark|first1=David P.|title=Molecular Biology|url=https://books.google.com.ar/books?id=m1SmtB4CiywC&pg=PT655&dq=%221983%22+%22mullis%22&hl=en&sa=X&ved=0ahUKEwiHmMC-0P3SAhXEjJAKHWV7CDsQ6AEIHjAB#v=onepage&q=%221983%22%20%22mullis%22&f=false|accessdate=30 March 2017}}</ref> || [[wikipedia:United States|United States]] |
|- | |- | ||
| − | | 1984 || Technology launch|| American molecular geneticist [[wikipedia:Charles Cantor|Charles Cantor]] and David C. Schwartz develop the [[wikipedia:pulsed field gel electrophoresis|pulsed field gel electrophoresis]] (PFGE) technique, which uses short pulses from perpendicular electrical fields to separate large pieces of DNA.<ref name="Milestones of DNA technologies"/><ref>{{cite web|title=Pulsed Field Gel Electrophoresis|url=http://www.bio-rad.com/es-ar/applications-technologies/pulsed-field-gel-electrophoresis|website=bio-rad.com|accessdate=28 March 2017}}</ref> || | + | | 1984 || Technology launch|| American molecular geneticist [[wikipedia:Charles Cantor|Charles Cantor]] and David C. Schwartz develop the [[wikipedia:pulsed field gel electrophoresis|pulsed field gel electrophoresis]] (PFGE) technique, which uses short pulses from perpendicular electrical fields to separate large pieces of DNA.<ref name="Milestones of DNA technologies"/><ref>{{cite web|title=Pulsed Field Gel Electrophoresis|url=http://www.bio-rad.com/es-ar/applications-technologies/pulsed-field-gel-electrophoresis|website=bio-rad.com|accessdate=28 March 2017}}</ref> || |
|- | |- | ||
| 1985 || Technology launch|| British geneticist [[wikipedia:Alec Jeffreys|Alec Jeffreys]] develops a method for DNA profiling. A DNA profile is produced by counting the number of short repeating sequences of DNA sequence found at ten specific regions of the genome.<ref>{{cite web|title=The history of genetic fingerprinting|url=http://www2.le.ac.uk/departments/genetics/jeffreys/history-gf|website=|publisher=University of Leicester|accessdate=30 March 2017}}</ref> || | | 1985 || Technology launch|| British geneticist [[wikipedia:Alec Jeffreys|Alec Jeffreys]] develops a method for DNA profiling. A DNA profile is produced by counting the number of short repeating sequences of DNA sequence found at ten specific regions of the genome.<ref>{{cite web|title=The history of genetic fingerprinting|url=http://www2.le.ac.uk/departments/genetics/jeffreys/history-gf|website=|publisher=University of Leicester|accessdate=30 March 2017}}</ref> || | ||
| Line 94: | Line 168: | ||
| 1990 || Organization || GATC Biotech is established as company specializing in DNA and RNA sequencing.<ref>{{cite web|title=GATC Biotech DNA Sequencing|url=https://www.nucleics.com/DNA_sequencing_support/sequencing-service/gatc-biotech.html|website=nucleics.com|accessdate=4 April 2017}}</ref> || [[wikipedia:Germany|Germany]] ([[wikipedia:Konstanz|Konstanz]]) | | 1990 || Organization || GATC Biotech is established as company specializing in DNA and RNA sequencing.<ref>{{cite web|title=GATC Biotech DNA Sequencing|url=https://www.nucleics.com/DNA_sequencing_support/sequencing-service/gatc-biotech.html|website=nucleics.com|accessdate=4 April 2017}}</ref> || [[wikipedia:Germany|Germany]] ([[wikipedia:Konstanz|Konstanz]]) | ||
|- | |- | ||
| − | | 1992 || Technology launch || The first "sequencing factory" is established.<ref name="DNA sequencing: bench to bedside and beyond"/> || | + | | 1992 || Technology launch || The first "sequencing factory" is established.<ref name="DNA sequencing: bench to bedside and beyond"/> || |
|- | |- | ||
| − | | 1993 || Organization || The Sanger Centre (today [[wikipedia:Wellcome Trust Sanger Institute|Wellcome Trust Sanger Institute) near Cambridge is opened by [[wikipedia:Frederick Sanger|Frederick Sanger]].<ref>{{cite book|last1=Wenkart|first1=Michael|title=50 scientific discoveries that changed the world|url=https://books.google.com.ar/books?id=jEG8AwAAQBAJ&pg=PA260&lpg=PA260&dq=%221993%22+%22sanger+centre%22&source=bl&ots=L78Q_v8NN7&sig=_CE-lnooNLtF5nKTY0mhAkV9_2k&hl=en&sa=X&ved=0ahUKEwiY-7fOuv_SAhXBEZAKHQ1lDeYQ6AEIRTAJ#v=onepage&q=%221993%22%20%22sanger%20centre%22&f=false|accessdate=31 March 2017}}</ref> || [[wikipedia:United Kingdom|United Kingdom]] ([[wikipedia:Hinxton|Hinxton]]) | + | | 1993 || Organization || The Sanger Centre (today [[wikipedia:Wellcome Trust Sanger Institute|Wellcome Trust Sanger Institute]]) near Cambridge is opened by [[wikipedia:Frederick Sanger|Frederick Sanger]].<ref>{{cite book|last1=Wenkart|first1=Michael|title=50 scientific discoveries that changed the world|url=https://books.google.com.ar/books?id=jEG8AwAAQBAJ&pg=PA260&lpg=PA260&dq=%221993%22+%22sanger+centre%22&source=bl&ots=L78Q_v8NN7&sig=_CE-lnooNLtF5nKTY0mhAkV9_2k&hl=en&sa=X&ved=0ahUKEwiY-7fOuv_SAhXBEZAKHQ1lDeYQ6AEIRTAJ#v=onepage&q=%221993%22%20%22sanger%20centre%22&f=false|accessdate=31 March 2017}}</ref> || [[wikipedia:United Kingdom|United Kingdom]] ([[wikipedia:Hinxton|Hinxton]]) |
|- | |- | ||
| − | | 1995 || Scientific development (non-human genome) || The first complete cellular genome sequences, from bacterium haemophilus influenzae by [[wikipedia:Craig Venter|Craig Venter]] and Nobel laureate [[wikipedia:Hamilton O. Smith|Hamilton O. Smith]] at the Institute of Genomic Research, appears. This would mark the first use of whole-genome shotgun sequencing, eliminating need of mapping efforts.<ref name="DNA sequencing: bench to bedside and beyond"/> || | + | | 1995 || Scientific development (non-human genome) || The first complete cellular genome sequences, from bacterium haemophilus influenzae by [[wikipedia:Craig Venter|Craig Venter]] and Nobel laureate [[wikipedia:Hamilton O. Smith|Hamilton O. Smith]] at the Institute of Genomic Research, appears. This would mark the first use of whole-genome shotgun sequencing, eliminating need of mapping efforts.<ref name="DNA sequencing: bench to bedside and beyond"/> || |
|- | |- | ||
| 1996 || Scientific development || [[wikipedia:Pål Nyrén|Pål Nyrén]] and [[wikipedia:Mostafa Ronaghi|Mostafa Ronaghi]], working at the Royal Institute of Technology, publish their Pyrosequencing method, based on sequencing by synthesis.<ref name="Biomarker Validation: Technological, Clinical and Commercial Aspects">{{cite book|title=Biomarker Validation: Technological, Clinical and Commercial Aspects|edition=Harald Seitz, Sarah Schumacher|url=https://books.google.com.ar/books?id=e5lxBgAAQBAJ&pg=PA191&lpg=PA191&dq=%22in+1996%22++%22Ronaghi+%22+%22nyren%22+%22Pyrosequencing+%22&source=bl&ots=eGmiEoXejx&sig=CCBApFqgxy2fvLrxXrH4arV-5LU&hl=en&sa=X&ved=0ahUKEwjN1rm_u__SAhVIS5AKHQHYDp4Q6AEIHTAC#v=onepage&q=%22in%201996%22%20%20%22Ronaghi%20%22%20%22nyren%22%20%22Pyrosequencing%20%22&f=false|accessdate=31 March 2017}}</ref> By 1998, pyrosequencing is introduced as first generation sequencing approach.<ref name="Books on Google Play Synthetic Biology and Metabolic Engineering in Plants and Microbes Part B: Metabolism in Plants"/> || [[wikipedia:Sweden|Sweden]] ([[wikipedia:Stockholm|Stockholm]]) | | 1996 || Scientific development || [[wikipedia:Pål Nyrén|Pål Nyrén]] and [[wikipedia:Mostafa Ronaghi|Mostafa Ronaghi]], working at the Royal Institute of Technology, publish their Pyrosequencing method, based on sequencing by synthesis.<ref name="Biomarker Validation: Technological, Clinical and Commercial Aspects">{{cite book|title=Biomarker Validation: Technological, Clinical and Commercial Aspects|edition=Harald Seitz, Sarah Schumacher|url=https://books.google.com.ar/books?id=e5lxBgAAQBAJ&pg=PA191&lpg=PA191&dq=%22in+1996%22++%22Ronaghi+%22+%22nyren%22+%22Pyrosequencing+%22&source=bl&ots=eGmiEoXejx&sig=CCBApFqgxy2fvLrxXrH4arV-5LU&hl=en&sa=X&ved=0ahUKEwjN1rm_u__SAhVIS5AKHQHYDp4Q6AEIHTAC#v=onepage&q=%22in%201996%22%20%20%22Ronaghi%20%22%20%22nyren%22%20%22Pyrosequencing%20%22&f=false|accessdate=31 March 2017}}</ref> By 1998, pyrosequencing is introduced as first generation sequencing approach.<ref name="Books on Google Play Synthetic Biology and Metabolic Engineering in Plants and Microbes Part B: Metabolism in Plants"/> || [[wikipedia:Sweden|Sweden]] ([[wikipedia:Stockholm|Stockholm]]) | ||
| Line 120: | Line 194: | ||
| 2000 || Technology launch|| University of California, Santa Cruz (UCSC) launches the [[wikipedia:UCSC Genome Browser|UCSC Genome Browser]], a graphical viewer for genomic data.<ref>{{cite journal|last1=Kuhn|first1=Robert M.|last2=Haussler|first2=David|last3=Kent|first3=W. James|title=The UCSC genome browser and associated tools|doi=10.1093/bib/bbs038|url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3603215/|accessdate=31 March 2017}}</ref> || | | 2000 || Technology launch|| University of California, Santa Cruz (UCSC) launches the [[wikipedia:UCSC Genome Browser|UCSC Genome Browser]], a graphical viewer for genomic data.<ref>{{cite journal|last1=Kuhn|first1=Robert M.|last2=Haussler|first2=David|last3=Kent|first3=W. James|title=The UCSC genome browser and associated tools|doi=10.1093/bib/bbs038|url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3603215/|accessdate=31 March 2017}}</ref> || | ||
|- | |- | ||
| − | | 2000 || Technology launch|| Lynx Therapeutics releases [[wikipedia:Massively parallel signature sequencing|Massively parallel signature sequencing]] (MPSS), a method that incorporates a parallelized, adapter/ligation-mediated, bead-based sequencing technology. || | + | | 2000 || Technology launch|| Lynx Therapeutics releases [[wikipedia:Massively parallel signature sequencing|Massively parallel signature sequencing]] (MPSS), a method that incorporates a parallelized, adapter/ligation-mediated, bead-based sequencing technology. || |
|- | |- | ||
| 2000 || Organization || [[wikipedia:454 Life Sciences|454 Life Sciences]] is founded as a biotechnology company specializing in sequencing of genetic samples. Acquired by [[wikipedia:Hoffmann-La Roche|Hoffmann-La Roche]] in 2007, it would be shut down in 2013 when its technology becomes noncompetitive.<ref>{{cite web|title=Roche to close 454 Life Sciences as it reduces gene sequencing focus|url=http://www.fiercebiotech.com/medical-devices/roche-to-close-454-life-sciences-as-it-reduces-gene-sequencing-focus|website=fiercebiotech.com|accessdate=4 April 2017}}</ref><ref>{{cite journal|last1=Patrick|first1=Kristin L.|title=454 Life Sciences: Illuminating the future of genome sequencing and personalized medicine|url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2347365/|accessdate=4 April 2017|pmc=2347365}}</ref> || [[wikipedia:United States|United States]] ([[wikipedia:|Branford, Connecticut]]) | | 2000 || Organization || [[wikipedia:454 Life Sciences|454 Life Sciences]] is founded as a biotechnology company specializing in sequencing of genetic samples. Acquired by [[wikipedia:Hoffmann-La Roche|Hoffmann-La Roche]] in 2007, it would be shut down in 2013 when its technology becomes noncompetitive.<ref>{{cite web|title=Roche to close 454 Life Sciences as it reduces gene sequencing focus|url=http://www.fiercebiotech.com/medical-devices/roche-to-close-454-life-sciences-as-it-reduces-gene-sequencing-focus|website=fiercebiotech.com|accessdate=4 April 2017}}</ref><ref>{{cite journal|last1=Patrick|first1=Kristin L.|title=454 Life Sciences: Illuminating the future of genome sequencing and personalized medicine|url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2347365/|accessdate=4 April 2017|pmc=2347365}}</ref> || [[wikipedia:United States|United States]] ([[wikipedia:|Branford, Connecticut]]) | ||
| Line 136: | Line 210: | ||
|- | |- | ||
| 2002 || Scientific development (non-human genome) || The genome of the parasite [[wikipedia:plasmodium falciparum|plasmodium falciparum]], which causes [[wikipedia:malaria|malaria]] in humans, is published.<ref>{{cite book|title=Advances in Malaria Research|edition=Deepak Gaur, Virander S. Chauhan, Chetan E. Chitnis|url=https://books.google.com.ar/books?id=p5NZDgAAQBAJ&pg=PA198&dq=%22in+2002%22+%09%22genome%22+%22plasmodium+falciparum%22&hl=en&sa=X&ved=0ahUKEwje1deq7__SAhXTl5AKHZTZA48Q6AEIGDAA#v=onepage&q=%22in%202002%22%20%09%22genome%22%20%22plasmodium%20falciparum%22&f=false|accessdate=31 March 2017}}</ref> || | | 2002 || Scientific development (non-human genome) || The genome of the parasite [[wikipedia:plasmodium falciparum|plasmodium falciparum]], which causes [[wikipedia:malaria|malaria]] in humans, is published.<ref>{{cite book|title=Advances in Malaria Research|edition=Deepak Gaur, Virander S. Chauhan, Chetan E. Chitnis|url=https://books.google.com.ar/books?id=p5NZDgAAQBAJ&pg=PA198&dq=%22in+2002%22+%09%22genome%22+%22plasmodium+falciparum%22&hl=en&sa=X&ved=0ahUKEwje1deq7__SAhXTl5AKHZTZA48Q6AEIGDAA#v=onepage&q=%22in%202002%22%20%09%22genome%22%20%22plasmodium%20falciparum%22&f=false|accessdate=31 March 2017}}</ref> || | ||
| − | |- | + | |- |
| − | | 2003 || Technology launch|| American geneticist [[wikipedia:George M. Church|George M. Church]], working at Harvard Medical School, | + | | 2003 || Technology launch|| American geneticist [[wikipedia:George M. Church|George M. Church]], working at Harvard Medical School, develops [[wikipedia:Polony sequencing|Polony sequencing]], multiplex sequencing technique that can be used to “read” millions of immobilized DNA sequences in parallel.<ref>{{cite web|title=Polony Sequencing|url=https://binf.snipcademy.com/lessons/ngs-techniques/polony-sequencing|website=snipcademy.com|accessdate=1 April 2017}}</ref> || [[wikipedia:United States|United States]] |
|- | |- | ||
| 2003 || Scientific development || Completion of the Human Genome Project is achieved. Aproximately 25,000 genes are identified. The human genome is sequenced to 99.99 per cent accuracy, 2 years ahead of schedule.<ref>{{cite book|title=Essentials of Medical Genetics for Health Professionals|url=https://books.google.com.ar/books?id=5FpM7VxEOqoC&pg=PA15&dq=%22in+2003%22+%22Human+Genome+Project%22+%22completed%22+%2225,000+genes%22&hl=en&sa=X&ved=0ahUKEwj11u_-8P_SAhWBF5AKHTLSAvoQ6AEINDAF#v=onepage&q=%22in%202003%22%20%22Human%20Genome%20Project%22%20%22completed%22%20%2225%2C000%20genes%22&f=false|accessdate=31 March 2017}}</ref><ref>{{cite book|title=Renaissance of Sickle Cell Disease Research in the Genome Era|edition=Betty S Pace|url=https://books.google.com.ar/books?id=32W7CgAAQBAJ&pg=PA31&dq=%222003%22+%22human+genome%22+%22ahead+of+schedule&hl=en&sa=X&ved=0ahUKEwjdspuN8v_SAhXGW5AKHa49DxcQ6AEILTAE#v=onepage&q=%222003%22%20%22human%20genome%22%20%22ahead%20of%20schedule&f=false|accessdate=31 March 2017}}</ref> || | | 2003 || Scientific development || Completion of the Human Genome Project is achieved. Aproximately 25,000 genes are identified. The human genome is sequenced to 99.99 per cent accuracy, 2 years ahead of schedule.<ref>{{cite book|title=Essentials of Medical Genetics for Health Professionals|url=https://books.google.com.ar/books?id=5FpM7VxEOqoC&pg=PA15&dq=%22in+2003%22+%22Human+Genome+Project%22+%22completed%22+%2225,000+genes%22&hl=en&sa=X&ved=0ahUKEwj11u_-8P_SAhWBF5AKHTLSAvoQ6AEINDAF#v=onepage&q=%22in%202003%22%20%22Human%20Genome%20Project%22%20%22completed%22%20%2225%2C000%20genes%22&f=false|accessdate=31 March 2017}}</ref><ref>{{cite book|title=Renaissance of Sickle Cell Disease Research in the Genome Era|edition=Betty S Pace|url=https://books.google.com.ar/books?id=32W7CgAAQBAJ&pg=PA31&dq=%222003%22+%22human+genome%22+%22ahead+of+schedule&hl=en&sa=X&ved=0ahUKEwjdspuN8v_SAhXGW5AKHa49DxcQ6AEILTAE#v=onepage&q=%222003%22%20%22human%20genome%22%20%22ahead%20of%20schedule&f=false|accessdate=31 March 2017}}</ref> || | ||
| Line 161: | Line 235: | ||
| 2008 || Program launch || The [[wikipedia:1000 Genomes Project|1000 Genomes Project]], an international research effort to establish by far the most detailed catalogue of human genetic variation, is launched. The project would finalize in 2012.<ref>{{cite book|last1=Hillert|first1=Dieter|title=The Nature of Language: Evolution, Paradigms and Circuits|url=https://books.google.com.ar/books?id=6owpBAAAQBAJ&pg=PA25&dq=%22in+2010%22+%09%221,000+Genomes+Project%22+%22Nature%22&hl=en&sa=X&ved=0ahUKEwjs_679gYDTAhVLf5AKHWcDCVkQ6AEIGDAA#v=onepage&q=%22in%202010%22%20%09%221%2C000%20Genomes%20Project%22%20%22Nature%22&f=false|accessdate=31 March 2017}}</ref>|| | | 2008 || Program launch || The [[wikipedia:1000 Genomes Project|1000 Genomes Project]], an international research effort to establish by far the most detailed catalogue of human genetic variation, is launched. The project would finalize in 2012.<ref>{{cite book|last1=Hillert|first1=Dieter|title=The Nature of Language: Evolution, Paradigms and Circuits|url=https://books.google.com.ar/books?id=6owpBAAAQBAJ&pg=PA25&dq=%22in+2010%22+%09%221,000+Genomes+Project%22+%22Nature%22&hl=en&sa=X&ved=0ahUKEwjs_679gYDTAhVLf5AKHWcDCVkQ6AEIGDAA#v=onepage&q=%22in%202010%22%20%09%221%2C000%20Genomes%20Project%22%20%22Nature%22&f=false|accessdate=31 March 2017}}</ref>|| | ||
|- | |- | ||
| − | | 2009 || Technology launch|| [[wikipedia:Pacific Biosciences|Pacific Biosciences]] introduces Single Molecule Real Time (SMRT) DNA Sequencing technique. It is based on the observation of the performance of polymerase during DNA synthesis.<ref>{{cite book|last1=Masoudi-Nejad|first1=Ali|last2=Narimani|first2=Zahra|last3=Hosseinkhan|first3=Nazanin|title=Next Generation Sequencing and Sequence Assembly: Methodologies and Algorithms|url=https://books.google.com.ar/books?id=8r28BAAAQBAJ&pg=PA25&dq=%22Nanopore+sequencing%22&hl=en&sa=X&ved=0ahUKEwjc8brqqILTAhVLf5AKHbkpDsIQ6AEIIDAC#v=onepage&q=%22Nanopore%20sequencing%22&f=false|accessdate=1 April 2017}}</ref> || | + | | 2009 || Technology launch|| [[wikipedia:Pacific Biosciences|Pacific Biosciences]] introduces Single Molecule Real Time (SMRT) DNA Sequencing technique. It is based on the observation of the performance of polymerase during DNA synthesis.<ref>{{cite book|last1=Masoudi-Nejad|first1=Ali|last2=Narimani|first2=Zahra|last3=Hosseinkhan|first3=Nazanin|title=Next Generation Sequencing and Sequence Assembly: Methodologies and Algorithms|url=https://books.google.com.ar/books?id=8r28BAAAQBAJ&pg=PA25&dq=%22Nanopore+sequencing%22&hl=en&sa=X&ved=0ahUKEwjc8brqqILTAhVLf5AKHbkpDsIQ6AEIIDAC#v=onepage&q=%22Nanopore%20sequencing%22&f=false|accessdate=1 April 2017}}</ref> || |
|- | |- | ||
| − | | 2009 || Scientific development || Researchers complete the sequence of the first cancer genomes from a [[wikipedia:lung cancer|lung tumour]] and a [[wikipedia:melanoma|melanoma.<ref>{{cite journal|editor1-link=Natascha Bushati, Bernard C Rossier|title=The Louis-Jeantet Prize 2013: Michael Stratton, Peter Hegemann and Georg Nagel|doi=10.1002/emmm.201202394|url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3569632/|accessdate=1 April 2017}}</ref> || | + | | 2009 || Scientific development || Researchers complete the sequence of the first cancer genomes from a [[wikipedia:lung cancer|lung tumour]] and a [[wikipedia:melanoma|melanoma]].<ref>{{cite journal|editor1-link=Natascha Bushati, Bernard C Rossier|title=The Louis-Jeantet Prize 2013: Michael Stratton, Peter Hegemann and Georg Nagel|doi=10.1002/emmm.201202394|url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3569632/|accessdate=1 April 2017}}</ref> || |
|- | |- | ||
| 2010 || Program launch || [[wikipedia:Wellcome Trust Sanger Institute|Wellcome Trust Sanger Institute]] launches UK10K project, which aims at studying a total of 10,000 human genomes in the United Kingdom, then compare the genomes of 4,000 healthy people with those of 6,000 people living with a disease of suspected genetic cause.<ref>{{cite web|title=How is the completed human genome sequence being used?|url=http://www.yourgenome.org/stories/how-is-the-completed-human-genome-sequence-being-used|website=yourgenome.org|accessdate=31 March 2017}}</ref> || | | 2010 || Program launch || [[wikipedia:Wellcome Trust Sanger Institute|Wellcome Trust Sanger Institute]] launches UK10K project, which aims at studying a total of 10,000 human genomes in the United Kingdom, then compare the genomes of 4,000 healthy people with those of 6,000 people living with a disease of suspected genetic cause.<ref>{{cite web|title=How is the completed human genome sequence being used?|url=http://www.yourgenome.org/stories/how-is-the-completed-human-genome-sequence-being-used|website=yourgenome.org|accessdate=31 March 2017}}</ref> || | ||
| Line 174: | Line 248: | ||
|- | |- | ||
| 2012 || Scientific development || ENCODE study publishes 30 research papers describing the active regions of the human genome including confirmation that the human genome contains 20,687 protein-coding genes.<ref name="Molecular and Cellular Regulation of Adaptation to Exercise"/><ref>{{cite web|title=ENCODE: the rough guide to the human genome|url=http://blogs.discovermagazine.com/notrocketscience/2012/09/05/encode-the-rough-guide-to-the-human-genome/#.WN3QPGekqUk|publisher=Discover Magazine|accessdate=31 March 2017}}</ref> || | | 2012 || Scientific development || ENCODE study publishes 30 research papers describing the active regions of the human genome including confirmation that the human genome contains 20,687 protein-coding genes.<ref name="Molecular and Cellular Regulation of Adaptation to Exercise"/><ref>{{cite web|title=ENCODE: the rough guide to the human genome|url=http://blogs.discovermagazine.com/notrocketscience/2012/09/05/encode-the-rough-guide-to-the-human-genome/#.WN3QPGekqUk|publisher=Discover Magazine|accessdate=31 March 2017}}</ref> || | ||
| − | |- | + | |- |
| 2013 || Policy || The United States Supreme Court rules that naturally occurring DNA cannot be patented, stating: "A naturally occurring DNA segment is a product of nature and not patent eligible merely because it has been isolated."<ref>{{cite web|title=Natural DNA Cannot Be Patented, Supreme Court Rules|url=http://www.npr.org/sections/thetwo-way/2013/06/13/191283148/natural-dna-cannot-be-patented-supreme-court-rules|website=npr.org|accessdate=29 March 2017}}</ref> || [[wikipedia:United States|United States]] | | 2013 || Policy || The United States Supreme Court rules that naturally occurring DNA cannot be patented, stating: "A naturally occurring DNA segment is a product of nature and not patent eligible merely because it has been isolated."<ref>{{cite web|title=Natural DNA Cannot Be Patented, Supreme Court Rules|url=http://www.npr.org/sections/thetwo-way/2013/06/13/191283148/natural-dna-cannot-be-patented-supreme-court-rules|website=npr.org|accessdate=29 March 2017}}</ref> || [[wikipedia:United States|United States]] | ||
|- | |- | ||
| Line 183: | Line 257: | ||
| 2014 || Organization || [[wikipedia:Genomics England|Genomics England]] is set up as company by the United Kingdom Department of Health to run the [[wikipedia:100,000 Genomes Project|100,000 Genomes Project]].<ref>{{cite web|title=Genomics England|url=https://www.wellcomegenomecampus.org/aboutus/genomicsengland.html|website=wellcomegenomecampus.org|accessdate=4 April 2017}}</ref> || [[wikipedia:United Kingdom|United Kingdom]] | | 2014 || Organization || [[wikipedia:Genomics England|Genomics England]] is set up as company by the United Kingdom Department of Health to run the [[wikipedia:100,000 Genomes Project|100,000 Genomes Project]].<ref>{{cite web|title=Genomics England|url=https://www.wellcomegenomecampus.org/aboutus/genomicsengland.html|website=wellcomegenomecampus.org|accessdate=4 April 2017}}</ref> || [[wikipedia:United Kingdom|United Kingdom]] | ||
|- | |- | ||
| − | | 2014 || || The average cost for a whole genome sequencing is less than US$ 10,000.<ref name="Biomarker Validation: Technological, Clinical and Commercial Aspects"/> || | + | | 2014 || || The average cost for a whole genome sequencing is less than US$ 10,000.<ref name="Biomarker Validation: Technological, Clinical and Commercial Aspects"/> || |
| + | |- | ||
| + | | 2015 (November) || Scientific development || International research team, using [[wikipedia:DNA sequencing|DNA sequencing]], manage to assess the extent to which geographical barriers in the [[wikipedia:Amazon rainforest|Amazon rainforest]] affect genetic variation among [[wikipedia:malaria|malaria]] vector ''[[wikipedia:Anopheles darlingi|Anopheles darlingi]]'' populations.<ref>{{cite web|title=DNA sequencing shows divergent genomes in malaria vectors of Brazilian rainforest|url=https://www.sciencedaily.com/releases/2015/11/151102083720.htm|website=sciencedaily.com|accessdate=25 June 2017}}</ref> || [[wikipedia:Amazon basin|Amazon basin]] | ||
|- | |- | ||
|} | |} | ||
| + | |||
| + | == See also == | ||
| + | |||
| + | * [[Timeline of CRISPR]] | ||
== References == | == References == | ||
| + | |||
| + | {{reflist|30em}} | ||
Latest revision as of 20:46, 4 June 2021
This is a timeline of DNA sequencing, focusing on scientific developments. Major discoveries concerning DNA are exposed as background information.
Contents
Big Picture
| Period | Key developments |
|---|---|
| 1930s–1940s | Molecular biology originates.[1] |
| 1950s–1960s | Period of extraordinary development in molecular biology, which becomes institutionalized. The structure of DNA is uncovered by James Watson and Francis Crick. Frederick Sanger plays a seminal role in the creation of influential DNA sequencing techniques in the 1950s and 1960s.[2][1] |
| 1970s | First-generation sequencing technologies merge, including the Maxam-Gilbert method, and the Sanger method.[3] Molecular biology goes genomic.[1] |
| 1980s–1990s | After the development of sequencing techniques, efforts are focused in sequencing the human genome, giving rise to the Human Genome Project. Scientists start sequencing DNA using capillary sequencers.[1][4] |
| 2000s | Second generation of DNA sequencing and RNA sequencing launches.[5] |
Numerical and visual data
Google Scholar
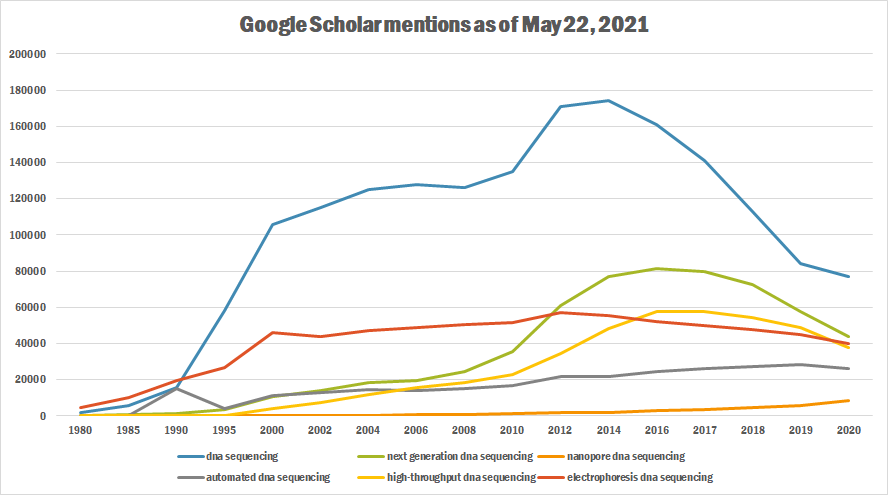
The following table summarizes per-year mentions on Google Scholar as of May 22, 2021.
| Year | dna sequencing | next generation dna sequencing | nanopore dna sequencing | automated dna sequencing | high-throughput dna sequencing | electrophoresis dna sequencing |
|---|---|---|---|---|---|---|
| 1980 | 1,660 | 164 | 100 | 11 | 4,710 | |
| 1985 | 5,880 | 475 | 1 | 361 | 20 | 10,300 |
| 1990 | 15,900 | 1,490 | 1,4980 | 66 | 19,400 | |
| 1995 | 58,000 | 3,340 | 5 | 4,320 | 369 | 26,800 |
| 2000 | 106,000 | 10,800 | 62 | 11,500 | 3,880 | 46,000 |
| 2002 | 115,000 | 14,100 | 129 | 12,900 | 7,290 | 44,000 |
| 2004 | 125,000 | 18,200 | 295 | 14,300 | 11,600 | 47,200 |
| 2006 | 128,000 | 19,800 | 477 | 14,200 | 15,700 | 48,900 |
| 2008 | 126,000 | 24,500 | 689 | 15,100 | 18,400 | 50,500 |
| 2010 | 135,000 | 35,300 | 1,140 | 16,600 | 23,100 | 51,800 |
| 2012 | 171,000 | 60,800 | 1,780 | 21,500 | 34,500 | 57,300 |
| 2014 | 174,000 | 76,900 | 2,100 | 21,900 | 48,500 | 55,700 |
| 2016 | 161,000 | 81,200 | 2,770 | 24,400 | 57,800 | 51,900 |
| 2017 | 141,000 | 79,800 | 3,630 | 25,900 | 57,500 | 49,700 |
| 2018 | 113,000 | 72,600 | 4,630 | 27,200 | 54,500 | 47,900 |
| 2019 | 84,000 | 57,600 | 5,790 | 28,600 | 48,800 | 44,800 |
| 2020 | 77,200 | 43,600 | 8,460 | 26,000 | 38,000 | 39,800 |
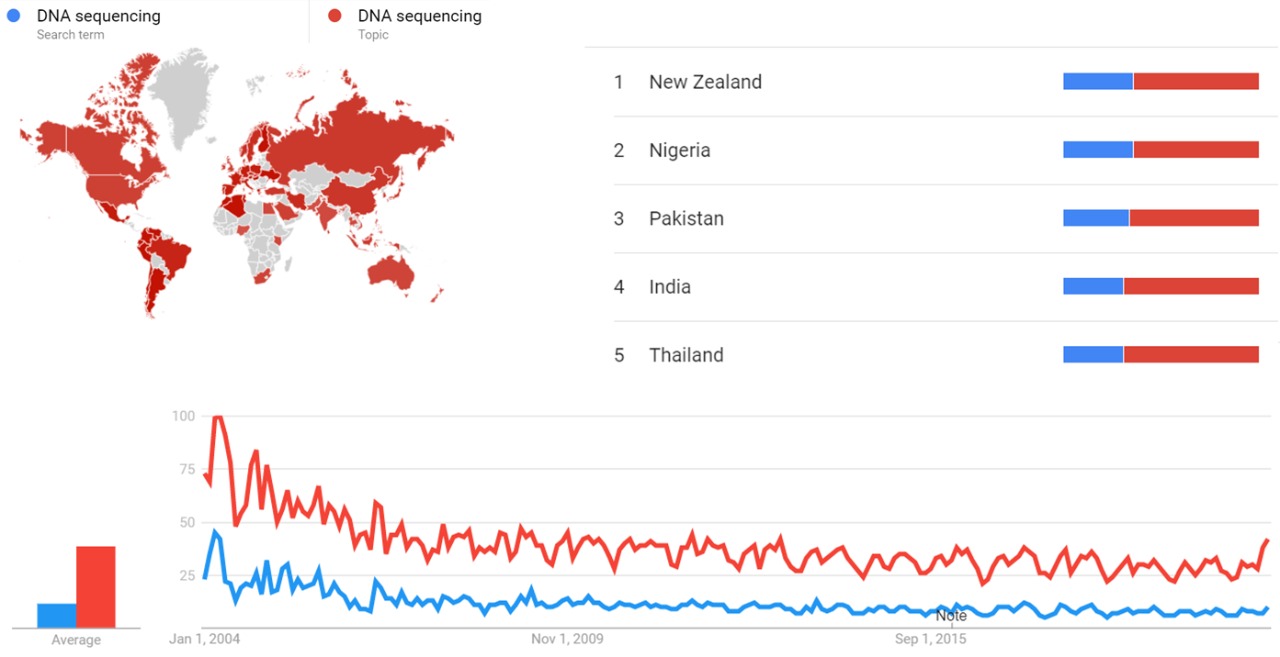
Google Trends
The comparative chart below shows Google Trends data for DNA sequencing (Search Term) and DNA sequencing (Topic), from January 2004 to February 2021, when the screenshot was taken. Interest in the topic is also ranked by country and displayed on world map.[6]
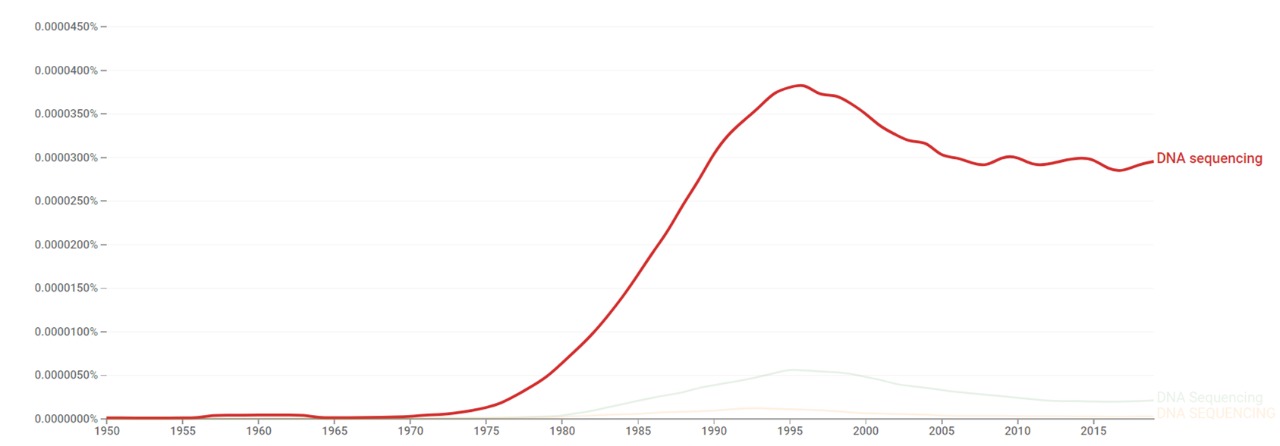
Google Ngram Viewer
The chart below shows Google Ngram Viewer data for DNA sequencing, from 1950 to 2019.[7]
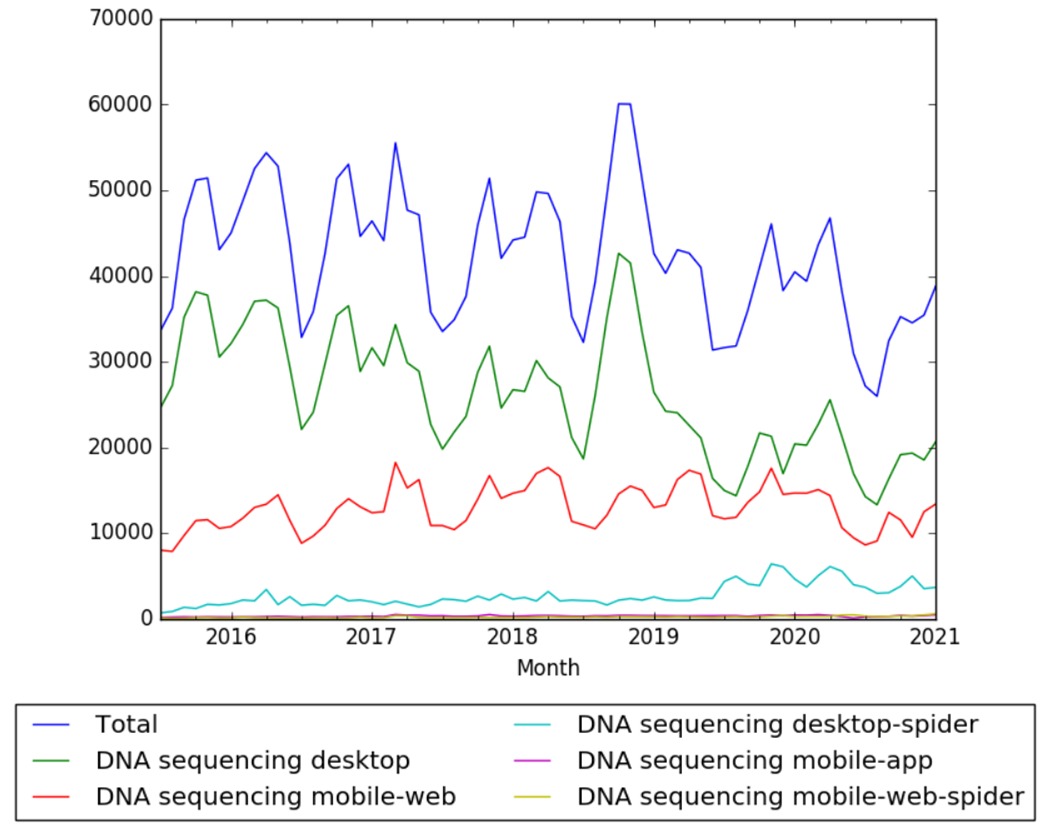
Wikipedia Views
The chart below shows pageviews of the English Wikipedia article DNA sequencing, on desktop, mobile-web, desktop-spider, mobile-web-spider and mobile app, from July 2015; to January 2021.[8]
Other
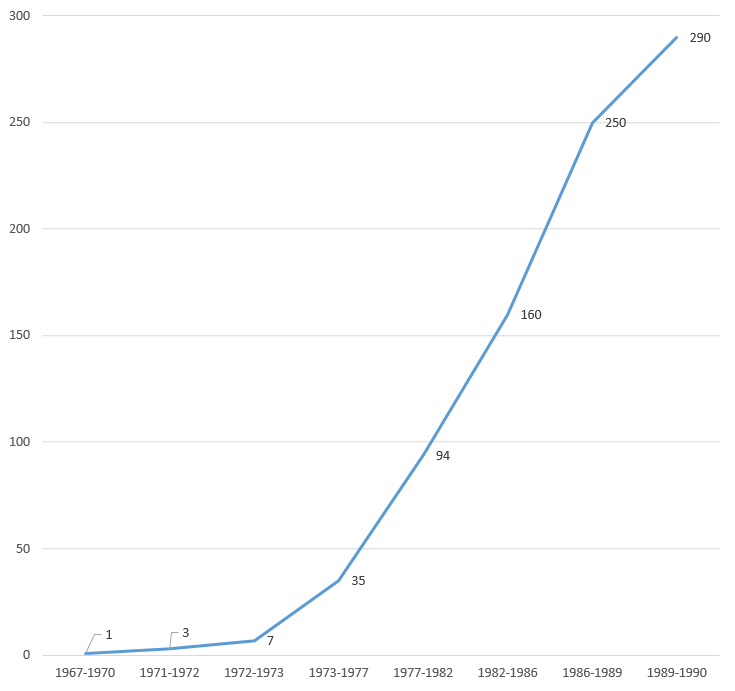
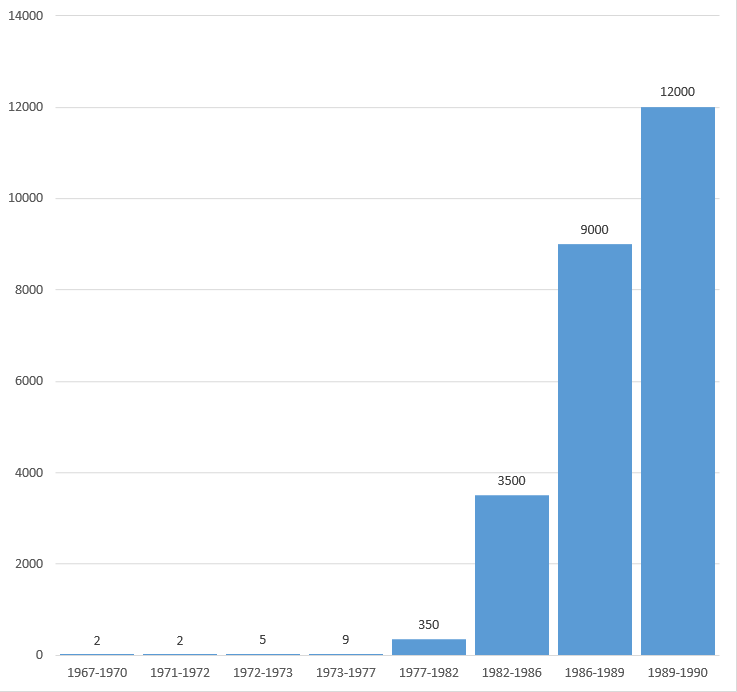
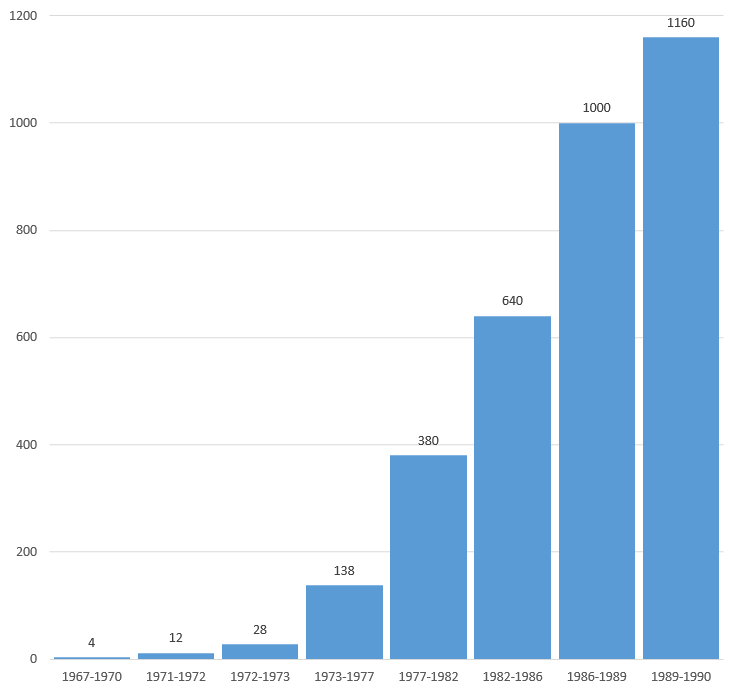
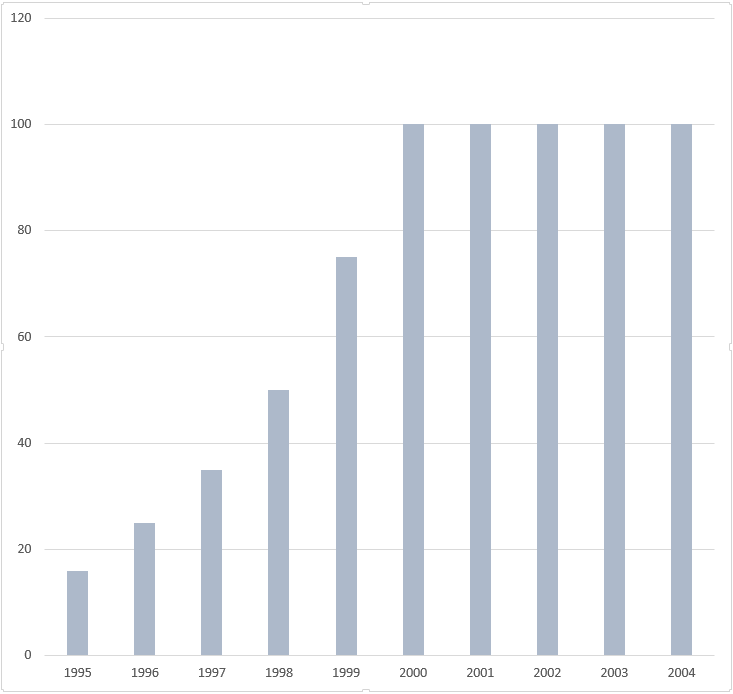
DNA sequence analysis between 1967 and 1990. Relative speed.[9]
Nucleotides determined per time period between 1967 and 1990.[9]
Number of investigators involved in DNA sequence analysis per time period between 1967 and 1990.[9]
DNA sequence analysis between 1967 and 1990. Nucleotides per year, per investigator.[9]
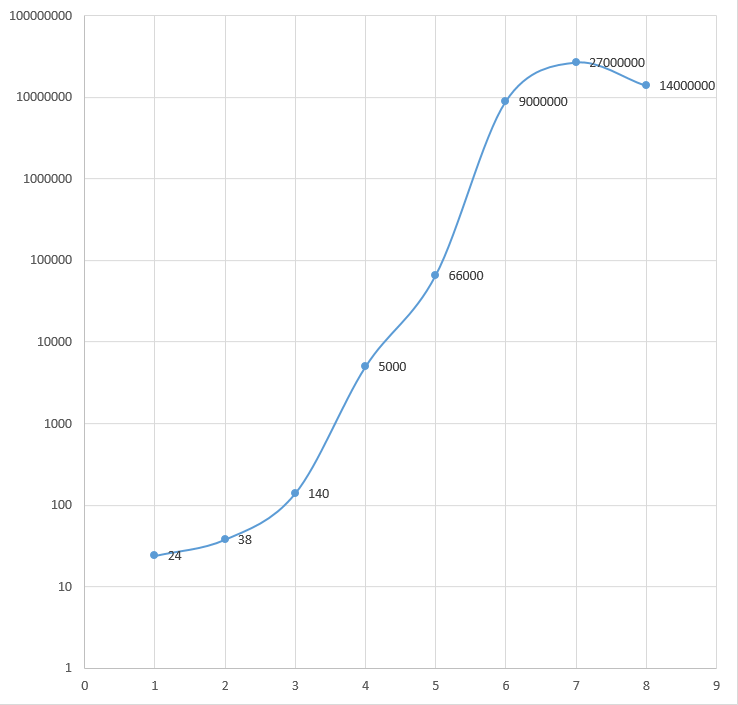
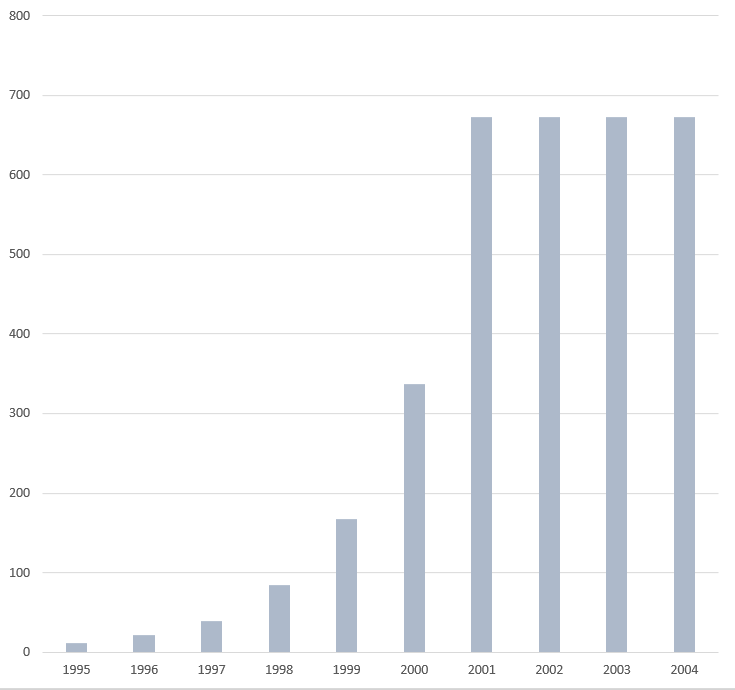
DNA sequencing costs in the Human Genome Project per year. Finished, per-base direct cost, in US$.[9]
DNA sequencing costs in the Human Genome Project. Annual sequencing budget (in US$ millions).[9]
DNA sequencing costs in the Human Genome Project. Genomic sequence (GB) per year, in US$ millions.[9]
DNA sequencing costs in the Human Genome Project per year. Genomic sequence (GB), cumulative, in US$ millions.[9]
Full timeline
| Year/period | Type of event | Event | Location |
|---|---|---|---|
| 1869–1871 | Scientific development | Swiss physician Friedrich Miescher first discovers and isolates Deoxyribonucleic acid (DNA). In 1871 Miescher publishes paper identifying the presence of nuclein (now known as DNA) and associated proteins, in the cell nucleus.[10] | Germany (Tübingen) |
| 1882 | Scientific development | German biochemist Albrecht Kossel differentiates ARN and DNA.[11] | |
| 1902 | Scientific development | American geneticist Walter Sutton publishes paper showing that chromosomes occur in distinct pairs, which segregate at meiosis.[12] | United States |
| 1902 | Scientific development | German biologist Theodor Boveri, working independently from Walter Sutton, recognizes that the behavior of Mendel’s particles during the production of gametes in peas precisely parallels the behavior of chromosomes at meiosis, suggesting that chromosomes could be shown to bear the material of heredity. This would further give rise to the so called Boveri–Sutton chromosome theory, which is recognized as a fundamental unifying theory of genetics which identifies chromosomes as the carriers of genetic material.[13] | Germany |
| 1906 | Scientific development | German biochemist Albrecht Kossel describes the four bases in nucleic acids (denine, cytosine, guanine, thymine). In 1910 Kossel would be awarded a Nobel Prize in Physiology or Medicine for that work.[11] | |
| 1944 | Scientific development | Canadian-American researchers Oswald Avery and Colin Munro MacLeod, along with American geneticist Maclyn McCarty, working on experiments on pneumococcal bacteria, establish that DNA could transform the properties of cells. As a result, a number of researchers would begin investigating the structure of DNA, hoping that this would reveal how the molecule works.[14] | |
| 1950 | Discovery | Austro-Hungarian biochemist Erwin Chargaff discovers that concentrations of thymine and adenine, and cytosine and guanine, are always found in equal amounts in samples of DNA. This suggests that A always pairs with T and C always pairs with G in a proportion that would later become known as Chargaff's ratio, an important clue for solving the structure of DNA. [15] | |
| 1952 | Scientific development | American bacteriologist Alfred Hershey and American geneticist Martha Chase conduct a series of experiments (today known as Hershey–Chase experiment) to demonstrate that DNA, rather than protein, carries our genetic information.[16] | |
| 1952 | Scientific development | R. Markham and J. D. Smith publish paper reporting that the hydrolysis of RNA proceeds via a cyclic phosphate intermediate, which is then further hydrolysed to produce a nucleoside 2'-monophosphate or 3'-monophosphate.[17] | |
| 1953 | Scientific development | American biologist James Watson and English physicist Francis Crick discover The double helix when solving the three-dimensional structure of DNA, working from crystallographic data produced by Rosalind Franklin and Maurice Wilkins.[18][19] Watson and Crick show that DNA has two strands made up of chemical sub-units known as nucleotides. These two strands arrange around each other, linked together by hydrogen bonds, in a spiral configuration called a double helix.[14] | |
| 1955 | Scientific development (non-human genome) | British biochemist Frederick Sanger completes the sequence of all the amino acids in the hormone insulin, a small protein secreted by the pancreas. For this achievement, Sanger would be awarded a Nobel Prize in Chemistry in 1958.[20] | |
| 1958 | Scientific development | English physicist Francis Crick publishes theory which argues that the arrangement of nucleotides in DNA determines the sequence of amino acids in proteins which in turn helps determine the function of a protein.[14] | |
| 1959 | Scientific development (non-human genome) | American molecular biologist Robert Sinsheimer reports the genome of bacteriophage Phi X 174 becoming the first DNA molecule purified to homogeneity.[18] | |
| 1961–1966 | Scientific development | American biochemist Marshall Warren Nirenberg, working at the National Institute of Arthritic and Metabolic Diseases, discover the first "triplet" —a sequence of three bases of DNA that codes for one of the twenty amino acids that serve as the building blocks of proteins. By 1966, Nirenberg has deciphered over sixty triplets. Nirenberg's experimental system would be used, confirmed and extended by Indian-American biochemist Har Gobind Khorana, who, along with Nirenberg and Robert W. Holley, was be awarded the 1968 Nobel Prize in Physiology or Medicine "for their interpretation of the genetic code and its function in protein synthesis."[21] | |
| 1965 | Scientific development (non-human genome) | Escherichia coli alanine transfer RNA becomes the first nucleic acid molecule to be sequenced by American biochemist Robert W. Holley and colleagues.[18][19] | |
| 1967 | Scientific development | U.E. Loening demonstrates that gels made from polymerized acrylamide and bisacrylamide ('polyacrylamide gels') have sufficient resolving power to separate high-molecular-weight pieces of RNA.[17] | |
| 1968 | Scientific development | Chinese-American biologist Ray Wu and Dale Kaiser publish the first successful DNA sequence achieved, detailing the sticky ends of the lambda phage. [22][18] [23] | |
| 1972 | Scientific development | American microbbiologist Hamilton O. Smith, at Johns Hopkins University School of Medicine, purifies the first site-specific "Type II" restriction enzyme, known as Hind II. Restriction enzymes would enable scientists to identify base sequences and, in combination with other tools, to manipulate DNA —and genes by the way— as never before. Hamilton Smith is awarded the Nobel Prize in physiology or medicine in 1978.[24] | United States |
| 1972 | Technology launch | Using the 2-D fractionation method developed by Frederick Sanger, Belgian molecular biologist Walter Fiers' laboratory produces the first complete protein-coding gene sequence, that of the coat protein of bacteriophage MS2.[19] | |
| 1973 | Scientific development | American molecular biologists Walter Gilbert and Allan Maxam publish first report of a nucleotide sequence with the lenght of 24 basepairs, using a method later known as wandering-spot analysis.[25][26][27] | |
| 1975 | Technology launch | Frederick Sanger introduces his ‘plus and minus’ method for DNA sequencing. This transition technique would prove to be critical, leading to the modern generation of methods that would completely dominate sequencing over the next 30 years.[18][28] | |
| 1976 | Scientific development (non-human genome) | Walter Fiers and coworkers at the University of Ghent identify and publish the sequence of the first complete genome of Bacteriophage MS2.[27] | |
| 1976–1977 | Technology launch | Walter Gilbert and Allan Maxam introduce their second method of DNA sequencing using chemical processes to terminate DNA strands. This method would be known as Maxam–Gilbert sequencing.[18][26] | |
| 1977 | Technology launch | Frederick Sanger develops his "chain-termination" or dideoxy technique, which allows the use of chemical analogues of the deoxyribonucleotides (dNTPs) that are the monomers of DNA strands. This technique is considered a major breakthrough, having altered the progress of DNA sequencing.[19] In 1980 Fred Sanger would be awarded his second Nobel Prize for Chemistry (this one along with Wally Gilbert and Paul Berg), for pioneering DNA sequencing methods.[20] | |
| 1981 | Scientific development (non-human genome) | The genome of the cauliflower mosaic virus is first to be sequenced by shotgun sequencing.[29] | |
| 1983 | Technology launch | American biochemist Kary Mullis develops the Polymerase chain reaction (PCR), a technique used in molecular biology to rapidly amplify DNA. The technique would prove revolutionary, and Mullis would be awarded the 1993 Nobel Prize in Chemistry. [30] | United States |
| 1984 | Technology launch | American molecular geneticist Charles Cantor and David C. Schwartz develop the pulsed field gel electrophoresis (PFGE) technique, which uses short pulses from perpendicular electrical fields to separate large pieces of DNA.[17][31] | |
| 1985 | Technology launch | British geneticist Alec Jeffreys develops a method for DNA profiling. A DNA profile is produced by counting the number of short repeating sequences of DNA sequence found at ten specific regions of the genome.[32] | |
| 1986 | Technology launch | Applied Biosystems Incorporated (API) markets the first automated DNA sequencer.[33] | |
| 1990 | Program launch | The Human Genome Project is launched, with aims at sequencing all 3 billion letters of a human genome in 15 years.[34] | |
| 1990 | Organization | GATC Biotech is established as company specializing in DNA and RNA sequencing.[35] | Germany (Konstanz) |
| 1992 | Technology launch | The first "sequencing factory" is established.[18] | |
| 1993 | Organization | The Sanger Centre (today Wellcome Trust Sanger Institute) near Cambridge is opened by Frederick Sanger.[36] | United Kingdom (Hinxton) |
| 1995 | Scientific development (non-human genome) | The first complete cellular genome sequences, from bacterium haemophilus influenzae by Craig Venter and Nobel laureate Hamilton O. Smith at the Institute of Genomic Research, appears. This would mark the first use of whole-genome shotgun sequencing, eliminating need of mapping efforts.[18] | |
| 1996 | Scientific development | Pål Nyrén and Mostafa Ronaghi, working at the Royal Institute of Technology, publish their Pyrosequencing method, based on sequencing by synthesis.[37] By 1998, pyrosequencing is introduced as first generation sequencing approach.[25] | Sweden (Stockholm) |
| 1996 | Scientific development (non-human genome) | An international effort of hundreds of researchers completes sequencing the genome of the budding yeast Saccharomyces cerevisiae, the first completely sequenced from a eukaryote.[38] | |
| 1996 | Policy | “Bermuda Principles” is formulated for Human Genome Project free data access.[39] | |
| 1997 | Organization | Macrogen is established as a public biotechnology company, specializing in DNA sequencing and bioinformatic analysis.[40] | South Korea (Seoul) |
| 1998 | Scientific development (non-human genome) | British biologist John Sulston and American biologist Bob Waterston publish the genome of the nematode worm, caenorhabditis elegans.[41] | |
| 1998 | Organization | American biotechnologist Craig Venter founds Celera Genomics Corporation for sequencing the human genome.[42] | |
| 1999 | Scientific development | The sequence of the euchromatic portion of human chromosome 22 is published. It is the first human chromosome to be sequenced as part of the Human Genome Project.[43] | |
| 1999 | Organization | Beijing Genomics Institute is founded. It is one of the world's genome sequencing centers.[44] | China (Shenzhen) |
| 2000 | Scientific development (non-human genome) | The full genome sequence of the model organism drosophila melanogaster (fruit fly) is completed by collaboration between the private company Celera Corporation and the public Berkeley Drosophila Genome Project (BDGP) based in California.[45] | |
| 2000 | Technology launch | University of California, Santa Cruz (UCSC) launches the UCSC Genome Browser, a graphical viewer for genomic data.[46] | |
| 2000 | Technology launch | Lynx Therapeutics releases Massively parallel signature sequencing (MPSS), a method that incorporates a parallelized, adapter/ligation-mediated, bead-based sequencing technology. | |
| 2000 | Organization | 454 Life Sciences is founded as a biotechnology company specializing in sequencing of genetic samples. Acquired by Hoffmann-La Roche in 2007, it would be shut down in 2013 when its technology becomes noncompetitive.[47][48] | United States (Branford, Connecticut) |
| 2001 | Scientific development | The first draft of the human genome sequence is released.[49] | |
| 2001 | Sequencing an entire genome is calculated to cost approximately US$ 100,000,000.[37] | ||
| 2002 | Scientific development (non-human genome) | The International Mouse Genome Sequencing Consortium completes full genome sequence of The mouse (the first in a mammal). The mouse genome is found to be 14 per cent smaller than the human genome, but over 95 per cent similar.[50] | |
| 2002 | Organization | Integromics is founded as a bioinformatics company. It provides products aimed at gene expression, sequencing and proteomics markets.[51] | Spain (Granada) |
| 2002 | Program launch | The International HapMap Project is launched to create a public database of human sequence variation.[52] | |
| 2002 | Scientific development (non-human genome) | The genome of the parasite plasmodium falciparum, which causes malaria in humans, is published.[53] | |
| 2003 | Technology launch | American geneticist George M. Church, working at Harvard Medical School, develops Polony sequencing, multiplex sequencing technique that can be used to “read” millions of immobilized DNA sequences in parallel.[54] | United States |
| 2003 | Scientific development | Completion of the Human Genome Project is achieved. Aproximately 25,000 genes are identified. The human genome is sequenced to 99.99 per cent accuracy, 2 years ahead of schedule.[55][56] | |
| 2003 | Program launch | The ENCODE (Encyclopedia of DNA Elements) project is launched by the National Human Genome Research Institute and aims at identifying and characterizing all the genes in the human genome.[57] | |
| 2004 | Organization | Pacific Biosciences, a company that develops and manufactures systems for gene sequencing, is founded.[58] | United States (Menlo Park, California) |
| 2005 | Technology launch | The total amount of sequence in the databases passes the 100 Gb mark.[18] | |
| 2005 | Organization | Oxford Nanopore Technologies, a company which develops nanopore sequencing products, is founded.[59] | United Kingdom |
| 2006 | Technology launch | American company Illumina next generation sequencing is on the market, after launching new Genoma Analizer (GA).[60] | United States |
| 2006 | Organization | Complete Genomics is founded as a life sciences company, specializing in DNA sequencing platform for human genome sequencing and analysis.[61] In 2013 Complete Genomics would be acquired by Beijing Genomics Institute.[62] | United States (Mountain View, California) |
| 2007 | Technology launch | Applied Biosystems releases new range of commercialized sequencers that implement a technology called Sequencing by Oligonucleotide Ligation Detection (SOLiD).[63] | |
| 2007 | Technology launch | A new DNA sequencing technology is introduced that increases DNA sequencing output 70 fold, in one year. This breakthrough would lead to the launch of the 1000 Genes Project in 2008.[64] | |
| 2007 | Organization | California-based company GenapSys starts selling sequencers.[65] | United States |
| 2008 | Program launch | The 1000 Genomes Project, an international research effort to establish by far the most detailed catalogue of human genetic variation, is launched. The project would finalize in 2012.[66] | |
| 2009 | Technology launch | Pacific Biosciences introduces Single Molecule Real Time (SMRT) DNA Sequencing technique. It is based on the observation of the performance of polymerase during DNA synthesis.[67] | |
| 2009 | Scientific development | Researchers complete the sequence of the first cancer genomes from a lung tumour and a melanoma.[68] | |
| 2010 | Program launch | Wellcome Trust Sanger Institute launches UK10K project, which aims at studying a total of 10,000 human genomes in the United Kingdom, then compare the genomes of 4,000 healthy people with those of 6,000 people living with a disease of suspected genetic cause.[69] | |
| 2010 | Scientific development | Nature magazine publishes the Neanderthal genome.[70] | |
| 2010 | Technology launch | American multinational Thermo Fisher Scientific launches the first lower-cost, nonoptical system which performs sequencing by synthesis, but with unlabeled nucleotides on a semiconductor chip. The full sequence is read by sequentially adding bases and tracking reactions across millions of microwells.[65] | United States |
| 2011 | Technology launch | Three major new sequencing platforms are released: Ion Torrent’s Personal Genome Machine (PGM), Pacific Biosciences’ RS and the Illumina MiSeq.[71] | |
| 2012 | Scientific development | ENCODE study publishes 30 research papers describing the active regions of the human genome including confirmation that the human genome contains 20,687 protein-coding genes.[57][72] | |
| 2013 | Policy | The United States Supreme Court rules that naturally occurring DNA cannot be patented, stating: "A naturally occurring DNA segment is a product of nature and not patent eligible merely because it has been isolated."[73] | United States |
| 2013 | Scientific development (non-human genome) | The Zebrafish genome sequence is completed.[74] | |
| 2014 | Technology launch | American company Illumina launches its HiSeq X Ten system, consisting of 10 ultra-high-throughput sequencers, each capable of generating up to 1.8 terabases of data in less than three days, with a sequencing capability of about 18,000 human genomes per year.[65] | United States |
| 2014 | Organization | Genomics England is set up as company by the United Kingdom Department of Health to run the 100,000 Genomes Project.[75] | United Kingdom |
| 2014 | The average cost for a whole genome sequencing is less than US$ 10,000.[37] | ||
| 2015 (November) | Scientific development | International research team, using DNA sequencing, manage to assess the extent to which geographical barriers in the Amazon rainforest affect genetic variation among malaria vector Anopheles darlingi populations.[76] | Amazon basin |
See also
References
- ↑ 1.0 1.1 1.2 1.3 "Molecular Biology". stanford.edu. Retrieved 1 April 2017.
- ↑ "Sequencing the worm". yourgenome.org. Retrieved 1 April 2017.
- ↑ "DNA sequencing". britannica.com. Retrieved 1 April 2017.
- ↑ "What is capillary sequencing?". yourgenome.org. Retrieved 1 April 2017.
- ↑ Biotechnology (Ulrich Kück, Nicole Frankenberg-Dinkel ed.). Retrieved 1 April 2017.
- ↑ "DNA sequencing". Google Trends. Retrieved 15 February 2021.
- ↑ "DNA sequencing". books.google.com. Retrieved 16 February 2021.
- ↑ "DNA sequencing". wikipediaviews.org. Retrieved 21 February 2021.
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 9.7 Cantor, Charles R.; Smith, Cassandra L. Genomics: The Science and Technology Behind the Human Genome Project. Retrieved 3 April 2017.
- ↑ "Friedrich Miescher and the discovery of DNA". sciencedirect.com. Retrieved 29 March 2017.
- ↑ 11.0 11.1 Vasudevan, D M; Sreekumari, S; Vaidyanathan, Kannan. Textbook of Biochemistry for Medical Students. Retrieved 29 March 2017.
- ↑ "100 Years Ago: Walter Sutton and the Chromosome Theory of Heredity". genetics.org. Retrieved 29 March 2017.
- ↑ "Historical development of the chromosome theory". An Introduction to Genetic Analysis. 7th edition. Retrieved 29 March 2017.
- ↑ 14.0 14.1 14.2 "The path to sequencing nucleic acids". whatisbiotechnology.org. Retrieved 29 March 2017.
- ↑ "Chargaff's Ratio". hhmi.org. Retrieved 29 March 2017.
- ↑ Saluz, Jost, H. DNA Methylation: Molecular Biology and Biological Significance. Retrieved 29 March 2017.
- ↑ 17.0 17.1 17.2 "Milestones of DNA technologies". Nature. doi:10.1038/nrg2250. Retrieved 28 March 2017.
- ↑ 18.0 18.1 18.2 18.3 18.4 18.5 18.6 18.7 18.8 Hutchison, Clyde A. "DNA sequencing: bench to bedside and beyond". PMC 2094077
 . doi:10.1093/nar/gkm688. Retrieved 28 March 2017.
. doi:10.1093/nar/gkm688. Retrieved 28 March 2017.
- ↑ 19.0 19.1 19.2 19.3 Heather, James M; Chain, Benjamin. "The sequence of sequencers: The history of sequencing DNA". sciencedirect.com. Retrieved 28 March 2017.
- ↑ 20.0 20.1 Roberts, Michael; Reiss, Michael Jonathan; Monger, Grace. Advanced Biology. Retrieved 29 March 2017.
- ↑ "Marshall Nirenberg (1927-) cracks the genetic code". genomenewsnetwork.org. Retrieved 30 March 2017.
- ↑ Sanger, Fred. "The early days of DNA sequences". doi:10.1038/85389. Retrieved 30 March 2017.
- ↑ "Ray Wu". whatisbiotechnology.org. Retrieved 30 March 2017.
- ↑ "Hamilton O. Smith (1931-) discovers the first site-specific restriction enzyme". genomenewsnetwork.org. Retrieved 30 March 2017.
- ↑ 25.0 25.1 Books on Google Play Synthetic Biology and Metabolic Engineering in Plants and Microbes Part B: Metabolism in Plants. Retrieved 30 March 2017.
- ↑ 26.0 26.1 "Maxam-Gilbert". snipcademy.com. Retrieved 30 March 2017.
- ↑ 27.0 27.1 Schiller, Jon. Genome Mapping: To Determine Disease Susceptibility. Retrieved 30 March 2017.
- ↑ Chan, EY. "Advances in sequencing technology.". doi:10.1016/j.mrfmmm.2005.01.004. Retrieved 28 March 2017.
- ↑ Gardner, Richard C.; Howarth, Alan J.; Hahn, Peter; Brown-Luedi, Marianne; Shepherd, Robert J.; Messing, Joachim (1981-06-25). "The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing". Nucleic Acids Research. 9 (12): 2871–2888. ISSN 0305-1048. PMC 326899
 . PMID 6269062. doi:10.1093/nar/9.12.2871. Retrieved 30 March 2017.
. PMID 6269062. doi:10.1093/nar/9.12.2871. Retrieved 30 March 2017.
- ↑ Clark, David P. Molecular Biology. Retrieved 30 March 2017.
- ↑ "Pulsed Field Gel Electrophoresis". bio-rad.com. Retrieved 28 March 2017.
- ↑ "The history of genetic fingerprinting". University of Leicester. Retrieved 30 March 2017.
- ↑ "Leroy Hood (1938-) develops the automated sequencer". genomenewsnetwork.org. Retrieved 30 March 2017.
- ↑ Chial, Heidi. "DNA Sequencing Technologies Key to the Human Genome Project". nature.com. Retrieved 30 March 2017.
- ↑ "GATC Biotech DNA Sequencing". nucleics.com. Retrieved 4 April 2017.
- ↑ Wenkart, Michael. 50 scientific discoveries that changed the world. Retrieved 31 March 2017.
- ↑ 37.0 37.1 37.2 Biomarker Validation: Technological, Clinical and Commercial Aspects (Harald Seitz, Sarah Schumacher ed.). Retrieved 31 March 2017.
- ↑ "The Reference Genome Sequence of Saccharomyces cerevisiae: Then and Now". doi:10.1534/g3.113.008995. Retrieved 31 March 2017.
- ↑ Powledge, Tabitha M. "Revisiting Bermuda". doi:10.1186/gb-spotlight-20030311-01. Retrieved 31 March 2017.
- ↑ "Box 2. Macrogen–a flagship for Korean biotechnology ventures". nature.com. Retrieved 4 April 2017.
- ↑ "A Transparent Window into Biology: A Primer on Caenorhabditis elegans". Genetics. Retrieved 31 March 2017.
- ↑ Müller-Wille, Staffan; Rheinberger, Hans-Jörg. A Cultural History of Heredity. Retrieved 30 March 2017.
- ↑ Pevsner, Jonathan. Bioinformatics and Functional Genomics. Retrieved 31 March 2017.
- ↑ "How Beijing Genomics Institute became the world's largest gene sequencer". foundingfuel.com. Retrieved 4 April 2017.
- ↑ Adams, Jill U. "Complex Genomes: Shotgun Sequencing". nature.com. Retrieved 31 March 2017.
- ↑ Kuhn, Robert M.; Haussler, David; Kent, W. James. "The UCSC genome browser and associated tools". doi:10.1093/bib/bbs038. Retrieved 31 March 2017.
- ↑ "Roche to close 454 Life Sciences as it reduces gene sequencing focus". fiercebiotech.com. Retrieved 4 April 2017.
- ↑ Patrick, Kristin L. "454 Life Sciences: Illuminating the future of genome sequencing and personalized medicine". PMC 2347365
 . Retrieved 4 April 2017.
. Retrieved 4 April 2017.
- ↑ Reuter, Jason A.; Spacek, Damek; Snyder, Michael P. "High-Throughput Sequencing Technologies". doi:10.1016/j.molcel.2015.05.004. Retrieved 31 March 2017.
- ↑ Sharma, Vinay. Text Book of Bioinformatics. Retrieved 31 March 2017.
- ↑ "Integromics S.L.". life-sciences-spain.com. Retrieved 4 April 2017.
- ↑ Genome Mapping and Genomics in Human and Non-Human Primates (Ravindranath Duggirala, Laura Almasy, Sarah Williams-Blangero, Solomon F.D. Paul, Chittaranjan Kole ed.). Retrieved 31 March 2017.
- ↑ Advances in Malaria Research (Deepak Gaur, Virander S. Chauhan, Chetan E. Chitnis ed.). Retrieved 31 March 2017.
- ↑ "Polony Sequencing". snipcademy.com. Retrieved 1 April 2017.
- ↑ Essentials of Medical Genetics for Health Professionals. Retrieved 31 March 2017.
- ↑ Renaissance of Sickle Cell Disease Research in the Genome Era (Betty S Pace ed.). Retrieved 31 March 2017.
- ↑ 57.0 57.1 Bouchard, Claude. Molecular and Cellular Regulation of Adaptation to Exercise. Retrieved 31 March 2017.
- ↑ "Pacific Biosciences' $600 Million Decoder Ring". bloomberg.com. Retrieved 1 April 2017.
- ↑ "Oxford Nanopore Technologies". crunchbase.com. Retrieved 1 April 2017.
- ↑ Tag-based Next Generation Sequencing (Matthias Harbers, Guenter Kahl, Günter Kahl ed.). Retrieved 31 March 2017.
- ↑ "Complete Genomics Sequences 14 Genomes". technologyreview.com. Retrieved 4 April 2017.
- ↑ "The Gene Factory". newyorker.com. Retrieved 4 April 2017.
- ↑ El-Metwally, Sara; Ouda, Osama M.; Helmy, Mohamed. Next Generation Sequencing Technologies and Challenges in Sequence Assembly. Retrieved 31 March 2017.
- ↑ "History of Genomics". news-medical.net. Retrieved 31 March 2017.
- ↑ 65.0 65.1 65.2 "Next-Gen Sequencing Is A Numbers Game". acs.org. Retrieved 4 April 2017.
- ↑ Hillert, Dieter. The Nature of Language: Evolution, Paradigms and Circuits. Retrieved 31 March 2017.
- ↑ Masoudi-Nejad, Ali; Narimani, Zahra; Hosseinkhan, Nazanin. Next Generation Sequencing and Sequence Assembly: Methodologies and Algorithms. Retrieved 1 April 2017.
- ↑ "The Louis-Jeantet Prize 2013: Michael Stratton, Peter Hegemann and Georg Nagel". doi:10.1002/emmm.201202394. Retrieved 1 April 2017.
- ↑ "How is the completed human genome sequence being used?". yourgenome.org. Retrieved 31 March 2017.
- ↑ "Ancient DNA reveals secrets of human history". nature.com. Retrieved 29 March 2017.
- ↑ "A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers". BMC Genomics. doi:10.1186/1471-2164-13-341. Retrieved 1 April 2017.
- ↑ "ENCODE: the rough guide to the human genome". Discover Magazine. Retrieved 31 March 2017.
- ↑ "Natural DNA Cannot Be Patented, Supreme Court Rules". npr.org. Retrieved 29 March 2017.
- ↑ The Zebrafish: Genetics, Genomics, and Transcriptomics. Retrieved 29 March 2017.
- ↑ "Genomics England". wellcomegenomecampus.org. Retrieved 4 April 2017.
- ↑ "DNA sequencing shows divergent genomes in malaria vectors of Brazilian rainforest". sciencedaily.com. Retrieved 25 June 2017.